Note: Descriptions are shown in the official language in which they were submitted.
TITLE: Methods and Kits for Monitoring Response to Radiation Therapies in
Cancer
[0001]
This is a Patent Cooperation Treaty Application which claims the benefit of 35
U.S.C.
119 based on the priority of U.S. Provisional Patent Application No.
61/732,697 filed December 3, 2012;
U.S. Provisional Patent Application No. 61/806,222 filed March 28, 2013 and
Patent Cooperation Treaty
Application No PCT/CA2013/000408 filed April 24, 2013 which are each
incorporated herein by reference
in their entirety.
Field
[0002]
The disclosure relates to methods and kits for evaluating response to
radiation in cancer
cells and tissues, and to monitoring response of subjects with cancer and
particularly breast and ovarian
cancer treated with modalities comprising radiation therapy.
Introduction
[0003]
PCT application (PCT/CA2008/001561) entitled "Method of Using Tumour RNA
Integrity
to Measure Response to Chemotherapy in Cancer Patients herein incorporated by
reference discloses a
method for monitoring response to chemotherapy in patients with locally
advanced breast cancer by
monitoring the ability of the chemotherapy agents to induce RNA degradation
(loss of RNA integrity), as
exhibited through a reduction in known metrics of RNA quality, including the
RNA integrity number
(RI N)52.
[0004] In
association with a national clinical trial (CAN-NCIC-CTG-MA.22), it was
demonstrated
that tumour RNA integrity number (RI N) values fell significantly upon
treatment of locally advanced breast
cancer patients with epirubicin/docetaxel chemotherapy and this response could
be significantly
correlated with the dose level of the regimen (p=0.05)53. Epirubicin, is an
epimer of doxorubicin, and both
compounds (known as anthracyclines) intercalate between DNA strands within
cells54. The drugs also
inhibit topoisomerase 1155 and DNA helicase56, thereby blocking DNA
replication. In addition, the drugs
are cytotoxic through the generation of free radicals, damaging a variety of
macromolecules including
DNA and lipids57. Docetaxel, in contrast, is an analog of paclitaxel. Both
drugs (known as taxanes) bind
to microtubules and prevent their dep01ymerisati0n58. This results in arrest
of cell cycle progression at
mitosis and mitotic catastrophe, and ultimately, the induction of apoptosism.
Unlike tumour extent
(cellularity) mid-treatment, it was observed in the MA.22 clinical trial that
low mid-treatment tumour RIN
values were predictive of pathologic complete response following treatment in
these patients (p=0.05)53.
1
7944618
Date Recue/Date Received 2022-10-28
summary
[0005] An
aspect includes a method of determining in a radiation treated cancer cell
sample, a
RNA profile comprising obtaining a cancer cell sample which has received one
or more doses of
radiation; isolating RNA from the sample, the isolated RNA comprising one or
more of radiation induced
degraded RNA (e.g disrupted RNA) and/or intact RNA, separating the isolated
RNA radiation induced
degraded RNA from intact RNA and measuring the amount of radiation induced
degraded RNA and/or
measuring the amount of intact RNA to provide a RNA profile of the radiation
treated cancer cell sample.
[0006] In
an embodiment, the measuring the amount of radiation induced degraded RNA
and/or
measuring the amount of intact RNA to provide a RNA profile of the radiation
treated cancer cell sample
comprises obtaining an electropherogram dataset from which a RNA profile is
determined. The profile
can comprise features from the electropherogram. In an embodiment, the RNA
profile is used to
determine an RNA integrity value. The RNA integrity value can be a composite
value determined from
different regions of the electropherogram.
[0007]
Another aspect includes a method of evaluating a cancer cell sample optionally
selected
from a breast cancer cell sample and an ovarian cancer cell sample, the method
comprising:
a. assaying the
cancer cell sample, optionally a breast cancer cell sample or
ovarian cancer cell sample, that has been exposed to a radiation dose to
obtain a RNA integrity value
and optionally to obtain a RNA concentration of the cancer cell sample;
b. identifying an increase, decrease or absence of change in the RNA
integrity
value and optionally an increase or decrease or absence of change in the RNA
concentration
compared to a control; and
c. optionally determining a radiation response score derived from the
increase,
decrease or absence of change in the RNA integrity value and optionally the
increase, decrease or
absence of change in the RNA concentration.
[0008] In
an embodiment, the increase or decrease in the RNA integrity value and
optionally the
increase or decrease or absence of change in the RNA concentration compared to
the control which is
used to calculate a radiation response score, indicates if the cancer cell
sample is responding to the
radiation dose.
[0009] In
an embodiment the method comprises the step of obtaining the cancer cell
sample,
optionally a breast cancer cell sample or ovarian cancer cell sample, from a
cancer cell source that has
been exposed to a radiation dose prior to assaying the cancer cell sample.
[00010] Another aspect provides a method of evaluating a cancer cell sample
optionally selected
from a breast cancer cell sample and an ovarian cancer cell sample, the method
comprising:
a.
obtaining the cancer cell sample optionally the breast cancer cell sample or
the
ovarian cancer cell sample from that has been exposed to a radiation dose;
2
7944618
Date Recue/Date Received 2022-10-28
b. assaying the
cancer cell sample to obtain a RNA integrity value and optionally
measure a RNA concentration of the cancer cell sample;
c.
comparing the RNA integrity value and optionally the RNA concentration to a
control; and
d.
identifying an increase, decrease or absence of change in the RNA integrity
value and optionally the RNA concentration compared to the control.
[00011] In an embodiment, the method further comprises determining a radiation
response score
derived from the increase, decrease or absence of change in the RNA integrity
value and optionally the
increase, decrease or absence of change in the RNA concentration.
[00012] A further aspect provides a method of evaluating a cancer cell sample
radiation
.. response, the method comprising:
a. obtaining a cancer cell sample, optionally a breast cancer cell sample
or an
ovarian cancer cell sample, that has been exposed to a radiation dose;
b. assaying the cancer cell sample to obtain a RNA integrity value and
optionally a
RNA concentration of the cancer cell sample;
c. comparing the RNA
integrity value and optionally the RNA concentration to a
control;
d.
identifying an increase, decrease or absence of change in the RNA integrity
value and optionally RNA concentration compared to the control; and
e. determining a radiation response score derived from the increase, decrease
or absence of
change in the RNA integrity value and optionally the RNA concentration
compared to the control.
[00013] In
an embodiment, the control is one or more predetermined reference values, for
example derived from a pretreatment sample or an earlier treatment sample or
derived from a group of
patients for example in a clinical trial. In an embodiment, the RNA integrity
value is proportional to the
degree of RNA degradation in the cancer sample. In an embodiment, the RNA
integrity value is inverse
proportional to the degree of RNA degradation in the cancer sample.
[00014]
The RNA integrity of RNA in a cancer cell sample can be measured for example
by
measuring a RNA integrity value. In an embodiment, the RNA integrity value is
proportional to the RNA
integrity of the sample, for example in such a case a high or increased RNA
integrity value corresponds
to a high or increased RNA integrity. In an embodiment, the method further
comprises comparing the
RNA integrity value and optionally the RNA concentration of the cancer cell
sample to a control such as
one or more response thresholds wherein a decreased RNA integrity value (e.g.
increased RNA
degradation) and optionally decreased RNA concentration of the cancer cell
sample compared to the
response threshold is predictive/indicative that the cancer cells or subject
are responsive to the radiation
therapy and a comparable or an increased RNA integrity value and optionally
increased concentration of
the cancer cell sample compared to the response threshold is
predictive/indicative the cancer cells are
unresponsive to the radiation treatment.
3
7944618
Date Recue/Date Received 2022-10-28
[00015] In an
embodiment, the comparing comprises comparing to a reference electropherogram
or a reference RNA integrity value derived therefrom, for example derived from
reference patients/cancer
cells treated with a reference radiation treatment. Different reference
electropherograms or reference
RNA integrity values correspond to different radiation treatments (e.g. with
or without a cytotoxic therapy
such as chemotherapy, the type of cytotoxic therapy, different doses,
different schedules etc).
[00016] In
embodiments wherein the RNA integrity value is inverse proportional, e.g.
measured
by RDI, a high RNA integrity value is indicative of degraded or low RNA
integrity, and a decreased RNA
integrity value compared to the control (optionally one or more response
thresholds) is
predictive/indicative that the cancer cells are (or the subject is)
unresponsive to the radiation treatment
and an increased RNA integrity value compared to the control (e.g. one or more
response thresholds) is
predictive/indicative that the cancer cells are (or the subject is) responsive
to the radiation treatment.
[00017] In
an embodiment, the cancer cells have (and/or the patient has) received 1, 2,
3, 4, 5, 6
or more doses of radiation.
[00018] In
another embodiment, the cancer cells are ( and/or the patient is) treated with
a dose of
a chemotherapeutic agent prior to and/or concurrent with radiation treatment.
[00019] In an embodiment, the chemotherapeutic agent is selected from taxane
chemotherapeutics, anthracyclines and combinations thereof.
[00020] In
a further embodiment, the taxane is selected from paclitaxel, docetaxel,
larotaxel,
AbraxaneTM, docoxahexaenoic acid-linked paclitaxel, paclitaxel polyglumex,
Ortataxel, GenexolTM,
liposomal-encapsulated paclitaxel, and paclitaxel in a Vitamin E emulsion.
[00021] In an
embodiment, the cancer cell sample is obtained, during for example after a
first
dose of radiation or post treatment.
[00022] In
another embodiment, the subject is administered a radiosensitizing or
radioprotecting
compound, for example prior to obtaining the cancer cell sample. For example
the subject can be
administered a radiosensitizing compound such as a taxane and/or a platinum
compound, optionally
systemically or remotely. Typically, the dose of the radiosensitizing agent
such as a taxane is reduced
compared to in the absence of radiation.
[00023] In
an embodiment, the cancer cells (e.g. the cancer source) is contacted with a
compound to optionally radiosensitize or radioprotect the cells or determine
if the compound is a
radiosensitizer or a radioprotector.
[00024] Accordingly
another aspect includes a method of determining if a cancer cell is
radiosensitive comprising treating a cell with a dose of radiation, optionally
in combination with a
radiosentizing agent for a sufficient time, isolating RNA, measuring a RNA
integrity value, and comparing
to a control, optionally untreated or treated with a dose of radiation alone
or a radiosensitizing agent
4
7944618
Date Recue/Date Received 2022-10-28
alone; wherein a decrease in RNA integrity is indicative that the cancer cell
is radiosensitive and a stable
or comparable to control RNA integrity is indicative that the cancer cell is
radioresistant.
[00025] The cancer cell can optionally be in vitro and/or in vivo.
[00026] In
an embodiment, the radiosensitizer is selected from oxygen increasing agents,
hypoxic cell sensitizers, halogenated pyrimidines, and/or bioreductive agents.
[00027] Another
aspect includes a method of determining if an agent is a radiosensitizing
agent
or a radioprotecting agent comprising:
a. incubating a cell source with a test agent;
b. exposing the cell source to a radiation dose;
c. assaying a sample of the cell source to obtain a RNA integrity value and
optionally RNA concentration of a sample of the cell source incubated with the
test agent;
d. comparing the RNA integrity value and optionally RNA concentration to a
control
reference sample e.g. corresponding to a sample incubated in the absence of
the test agent and
exposed to the radiation dose; and
e. identifying the test agent as a radiosensitizing agent if the RNA
integrity value
indicates that that RNA integrity and optionally the RNA concentration is
decreased compared to the
control reference sample value and identifying the agent as a radioprotecting
agent if the RNA
integrity value indicates that that RNA integrity and optionally the RNA
concentration is increased
compared to the control reference sample.
[00028] In
an embodiment, the method comprises determining a radiation response score
derived from the increase, decrease or absence of change in the RNA integrity
value and optionally the
RNA concentration compared to the control. The radiation response score can
for example be used to
rank or group the radioprotecting or radiosensitizing effectiveness of various
test agents.
[00029] In
an embodiment the cell source is incubated with (or a non-human animal is
administered) the test agent for at least 24h, at least 36h, at least 48 h, at
least 60h or at least 72 h
before measuring the RNA integrity and/or RNA concentration of the sample.
[00030]
The test agent can also in an embodiment be administered to a subject having
or
suspected of having a cancer. For example, the test agent can be a
radiosensitizing agent such as a
taxane that is administered to the subject prior to radiation to determine
tumour sensitivity to radiation.
[00031] In an embodiment, the cell used with the test agent is a cancer
cell.
[00032] In another
embodiment, the cancer cells are exposed to the radiation dose in vitro or in
vivo. For example, cancer cells exposed in vivo includes where the subject is
exposed to the radiation
dose and/or receives the radiation comprising treatment regimen.
[00033] In
an embodiment the control is a response threshold or one or more response
thresholds.
5
7944618
Date Recue/Date Received 2022-10-28
[00034] In another embodiment, the response threshold is derived from an
untreated sample
and/or is a standard derived from a plurality of untreated samples.
[00035] In yet another embodiment, the response threshold is derived
from an unresponsive
sample and/or a responsive sample and/or is a standard derived from a
plurality of unresponsive
samples and/or responsive samples.
[00036] A further aspect includes a method for evaluating a patient-derived
cancer cell sample
optionally selected from a breast cancer cell sample or an ovarian cancer cell
sample, the method
comprising:
a. assaying the cancer cell sample obtained from a subject after the subject
has been
exposed to a radiation dose with or without pre-treatment or concurrent
treatment with a chemotherapy
agent to obtain a RNA integrity value and optionally a RNA concentration of
the cancer cell sample;
b. identifying an increase, or decrease or absence of change in the RNA
integrity value
and optionally an increase, decrease or absence of change in the RNA
concentration compared to a
control; and
c. optionally determining a response score derived from the increase or
decrease or
absence of change in the RNA integrity value and optionally the increase or
decrease in the RNA
concentration.
[00037] In an embodiment, the method for evaluating a patient-derived
cancer cell sample
optionally selected from a breast cancer cell sample or an ovarian cancer cell
sample, the method
comprising:
a. obtaining a cancer cell sample, optionally a breast cancer cell sample or
an ovarian
cancer cell sample from a subject after the subject has been exposed to a
radiation dose with or without
pre-treatment or concurrent treatment with a cytotoxic, optionally
chemotherapy agent; and
b. assaying the cancer cell sample to obtain a RNA integrity value and
optionally a
RNA concentration of the cancer cell sample.
[00038] In an embodiment, the method further comprises comparing the RNA
integrity value
and/or the RNA concentration of the patient cancer cell sample to a control
such as a response
threshold wherein a decreased RNA integrity indicated by the RNA integrity
value and/or a decreased
RNA concentration of the patient cancer cell sample compared to the response
threshold is predictive
that the patient is responsive to the radiation therapy and a comparable (e.g.
absence of change) or an
increased RNA integrity indicated by the RNA integrity value and/or increased
RNA concentration of the
patient cancer cell sample compared to the response threshold is
predictive/indicative the patient is
unresponsive to the radiation treatment
[00039] Another aspect includes a method of identifying non-responding
patients comprising:
a. obtaining a cancer cell sample from the patient after the patient has been
exposed to one or more radiation doses;
6
7944618
Date Recue/Date Received 2022-10-28
b. assaying the cancer cell sample to obtain a RNA integrity value and/or a
RNA
concentration of the cancer cell sample;
c. comparing to a response threshold; and
d. identifying patients having a RNA integrity indicated by a RNA integrity
value
and/or a RNA concentration that is above the response threshold.
[00040] Yet a further aspect include, a method of predicting a treatment
outcome of a patient
having cancer optionally breast cancer or ovarian cancer, the method
comprising: assaying a cancer cell
sample obtained from the subject to obtain a RNA integrity value and/or RNA
concentration , wherein the
subject has been treated with a radiation dose, wherein a RNA integrity
indicated by the RNA integrity
value that is below a response threshold predicts subject/cancer response to
the radiation treatment and
a decreased risk of progression; and a RNA integrity indicated by the RNA
integrity value and/or RNA
concentration that is higher than the response threshold predicts
subject/cancer resistance to the
radiation treatment and an increased risk of disease progression.
[00041] Obtaining a RNA integrity value and/or RNA concentration or
obtaining a RNA integrity
value and optionally RNA concentration means that a RNA integrity value is
obtained alone or in
combination with a RNA concentration.
[00042] In an embodiment, if the subject/cancer is predicted to have an
increased risk of
progression, the method further comprises changing the treatment and if the
subject/cancer is predicted
to have a decreased risk of progression, continuing the treatment.
[00043] In an embodiment, the radiation dose and/or chemotherapy is
administered
preoperatively.
[00044] A further aspect includes a treatment method comprising:
a) exposing a patient with cancer to a radiation dose;
b) obtaining a cancer cell sample from the patient after administration of the
radiation dose;
c) assaying the cancer cell sample to obtain a RNA integrity value and
optionally a RNA
concentration of the cancer cell sample and
d) continuing the treatment when the RNA integrity indicated by the RNA
integrity value and
optionally the RNA concentration is decreased compared to a control such as a
response threshold and
changing the treatment when the RNA integrity indicated by the RNA integrity
value and optionally the
RNA concentration is comparable (e.g. absence of change) or increased compared
to the control,
optionally response threshold.
Also is provided in another aspect is use a method described herein for
treating a subject with
cancer, the use comprising a) assaying a cancer cell sample to obtain a RNA
integrity value and
optionally a RNA concentration of the cancer cell sample, wherein the patient
has been exposed to a
radiation dose and b) continuing to treat the patient when the RNA integrity
indicated by the RNA
integrity value and optionally the RNA concentration is decreased compared to
a control such as a
response threshold and changing the treatment when the RNA integrity indicated
by the RNA integrity
7
7944618
Date Recue/Date Received 2022-10-28
value and optionally the RNA concentration is comparable (e.g. absence of
change) or increased
compared to the control, optionally response threshold.
[00045] In an embodimentõ step b comprises assigning the patient to
continue the treatment
when the RNA integrity indicated by the RNA integrity value and optionally the
RNA concentration is
decreased compared to a control such as a response threshold and assigning the
patient to change the
treatment when the RNA integrity indicated by the RNA integrity value and
optionally the RNA
concentration is comparable (e.g. absence of change) or increased compared to
the control, optionally
response threshold.
[00046] In an embodiment, the cancer cell sample is a breast cancer
cell sample obtained from a
breast cancer patient.
[00047] In another embodiment, the breast cancer is of the Her2, basal,
lumina! A, luminal B, or
"normal" subtypes.
[00048] In another embodiment, the breast cancer patient has advanced
disease, including
locally advanced or inflammatory breast cancer (LABC).
[00049] In another embodiment, the cancer cell sample is an ovarian
cancer cell sample
obtained from an ovarian cancer patient.
[00050] In yet another embodiment, the response threshold is derived
from an untreated cancer,
optionally breast cancer or ovarian cancer patient or a standard derived from
a plurality of untreated
cancer, optionally breast cancer or ovarian cancer, patients.
[00051] In another embodiment, the response threshold derived untreated
cancer patient or the
standard derived from a plurality of untreated cancer patients is derived from
one or more pretreatment
samples.
[00052] In an embodiment, the response threshold predictive of
responsiveness is RNA integrity
and/or concentration decreased by at least 10%, at least 20%, at least 30%, at
least 40%, at least 50%,
at least 60%, at least 70%, at least 80% or at least 90% below a untreated
control such as a pretreatment
value.
[00053] In yet another embodiment, the response threshold is derived
from an unresponsive
patient cancer cell sample and/or a responsive patient cancer cell sample
and/or is a standard derived
from a plurality of unresponsive patient cancer cell samples and/or a
plurality of responsive patient
cancer cell samples.
[00054] In an embodiment, the patient is further treated with a dose of a
chemotherapeutic agent.
[00055] In a further embodiment, the chemotherapeutic agent is selected
from taxane
chemotherapeutics for example paclitaxel, docetaxel, larotaxel, AbraxaneTM,
docoxahexaenoic acid-
linked paclitaxel, paclitaxel polyglumex, Ortataxel, Genexorm, liposomal-
encapsulated paclitaxel, and
paclitaxel in a Vitamin E emulsion
8
7944618
Date Recue/Date Received 2022-10-28
[00056] In an embodiment, the taxane is docetaxel or paclitaxel.
[00057] In another embodiment, the cancer cell sample is obtained after
the patient has received
1, 2, 3, 4, 5, 6, or more doses of radiation treatment.
[00058] In another embodiment, two or more cancer cell samples are
obtained.
[00059] In another embodiment, the RNA integrity is measured by
electrophoretic separation of
RNA, optionally total RNA or ribosomal RNA.
[00060] In yet another embodiment, measuring the RNA integrity value
comprises calculating a
28S: 18S ribosomal (rRNA) ratio.
[00061] In an embodiment, the RNA integrity is measured by calculating
a RIN e.g., the RNA
integrity value is the RNA integrity number (RIN) and is assayed for example
using an Agilent bioanalyzer
machine. The RIN is calculated using an algorithm described in, Schroeder, A.,
0. Mueller, et al. 2006;
Mueller 2004; Vespucci 2005, e.g. Agilent Expert, which is used in the Agilent
2100 Bioanalyzer. The
method automatically selects features from signal measurements and constructs
regression models
based on a Bayesian learning technique. Feature spaces of different
dimensionality are compared in the
Bayesian framework, which allows selecting a final feature combination
corresponding to models with
high posterior probability. The approach was applied to a large collection of
electrophoretic RNA
measurements recorded with an Agilent 2100 bioanalyzer to develop an algorithm
that describes RNA
integrity. The resulting algorithm is a user-independent, automated and
reliable procedure for
standardization of RNA quality control that allows the calculation of an RNA
integrity number (RI N) under
certain conditions and/or for certain samples.
[00062] In yet another embodiment, the RNA integrity is measured using an
RNA disruption
assay as described in W02013/159200, incorporated herein in its entirety. The
RNA integrity value is in
an embodiment, a RNA disruption index (RDI) value. In another embodiment,
wherein the sample is a
patient sample, the RNA integrity value comprises a RDA zone.
[00063] In a further embodiment, RNA is isolated from the cancer cell
sample and the RNA
integrity value and/or the RNA concentration is measured on the isolated RNA.
[00064] In another embodiment, the RNA integrity value is measured by
separating the RNA by
microcapillary electrophoresis, and detecting RNA integrity with fluorescent
dyes.
[00065] In another embodiment, the RNA concentration is measured.
[00066] In an embodiment, the RNA integrity value is measured.
[00067] In another embodiment, the RNA integrity value is RNA integrity
number (RIN). In
another embodiment, the RNA integrity value is a RNA disruption assay value
e.g. a RDI or a RDA zone.
RDI can be more sensitive than RIN in measuring radiation induced effects. RDI
more accurately
captures cytotoxic e.g. radiation and/or chemotherapy induced RNA degradation.
9
7944618
Date Recue/Date Received 2022-10-28
[00068] In another
embodiment, both the RNA concentration and the RNA integrity are
measured.
[00069] In
yet another aspect is provided a method for remotely interpreting RNA
integrity of a
cancer cell sample, the method comprising:
a.
obtaining a cancer cell sample optionally selected from breast cancer and
ovarian cancer from a patient that has received a radiation dose at a remote
site;
b.
assaying the cancer cell sample to obtain one or more datasets selected from
an
electropherogram dataset, a RNA integrity value and optionally a RNA
concentration at the remote
site;
c. transmitting one or more datasets to a central site, by
internet;
d. generating, at
the central site, one or more interpretive data sets from the one or
more datasets using a computer;
e. i. generating, using a computer, one or more test reports under the
control of
one or more expert reviewers; and,
f. transmitting the one or more test reports to the remote site or another
site by
Internet.
[00070] In
yet another aspect is provided a method for remotely interpreting RNA
integrity of a
cancer cell sample, the method comprising:
a.
obtaining a cancer cell sample optionally selected from breast cancer and
ovarian cancer from a patient that has received a radiation dose at a remote
site;
b. sending the cancer cell sample to the central site:
c.
assaying the received cancer cell sample to obtain one or more datasets
selected from an electropherogram dataset, a RNA integrity value and
optionally a RNA concentration
at the central site;
d.
generating, at the central site, one or more interpretive data sets from the
one or
more datasets using a computer;
e.
generating, using a computer, one or more test reports under the control of
one
or more expert reviewers; and,
f.
transmitting the one or more test reports to the remote site or another site
by
internet.
[00071] The one or
more datasets including the electropherogram dataset and the one or
more interpretive datasets may be obtained and sent, for example, via a data
communication network or
wireless communication.
[00072] In
an embodiment, the cancer cell sample, for example obtained from the
subject, is placed in an RNA isolating or stabilizing composition prior to
assaying.
[00073] A further
aspect includes a kit for use in a method described herein comprising a
RNA isolating composition and an RNAse free vessel for receiving the cancer
cell sample and/or RNA
7944618
Date Recue/Date Received 2022-10-28
sample, wherein the vessel is optionally labeled with an identifier optionally
permitting for anonymous
testing.
[00074]
Other features and advantages of the present disclosure will become apparent
from the
following detailed description. It should be understood, however, that the
detailed description and the
specific examples while indicating preferred embodiments of the disclosure are
given by way of
illustration only, since various changes and modifications within the spirit
and scope of the disclosure will
become apparent to those skilled in the art from this detailed description.
Brief description of the drawinos
An embodiment of the present disclosure will now be described in relation to
the
drawings in which:
Figure 1: Schema of clinical trial conducted at the London Regional Health
Sciences
Centre, whereby patients with locally advanced breast cancer were treated
firstly by a chemotherapy
regimen including 5'Fuorouracil, epirubicin, and cyclophosphamide (FEC)
followed, by weekly docetaxel
with concurrent daily radiation, followed by weekly docetaxel.
Figure 2: Effect of FEC treatment and/or docetaxel treatment with concurrent
radiation
for patients in the London Clinical Trial depicted in Figure 1. Mean tumour
RNA concentrations
standard error at the various treatment times are depicted in the top panel,
while the bottom panel
depicts the biopsy RNA concentration values for each patient.
Figure 3: Inability of tumour RNA concentration values pre-, mid-, or post-
treatment by
themselves to indicate degree of clinical response in the London clinical
trial, which included a post-
treatment pathologic complete response (pCR), a partial response (PR), or no
response (NR), including
the presence of stable or progressive (prog) disease.
Figure 4: Graph of RIN values vs. RNA concentration for patients in the London
clinical
trial who achieved pCR (Responders) and patients who did not achieve pCR (Non-
Responders). In
Figure 4A, tumour samples were taken at mid-treatment (after FEC); one
Responder sample out of three
had a low RNA concentration and a low RIN value. In Figure 4B, tumour samples
were taken after
radiation and docetaxel; both Responder samples had low RNA concentrations and
low RIN values.
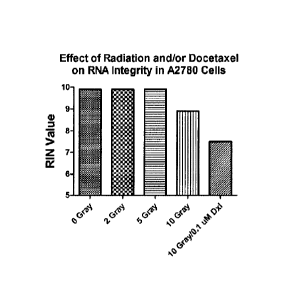
Figure 5: Effect of various doses of radiation on RNA content (concentration)
(panel A) or
RNA integrity (panel B) in A2780 ovarian tumour cells. The effect of 0.1 1.1M
docetaxel treatment with
concurrent 10 Gray radiation is also depicted. Radiation treatment alone
reduced RNA content and
integrity in an apparent dose-dependent manner, which was strongly augmented
by concurrent 0.1 iaM
docetaxel treatment.
Figure 6: Effect of various doses of radiation on RNA content (concentration)
(panel A) or
RNA integrity (panel B) in Her2+ SKBR3 breast tumour cells. The effect of 0.1
OA docetaxel treatment
11
7944618
Date Recue/Date Received 2022-10-28
with concurrent 10 Gray radiation is also depicted. Ten gray radiation
treatment alone slightly reduced
RNA content, with no apparent effect on RNA integrity. Effects of 10 gray
radiation on RNA content and
RNA integrity was strongly augmented by concurrent 0.1 1.1M docetaxel
treatment, in particular for RNA
integrity.
Figure 7: Effect of various doses of radiation on RNA content (concentration)
(panel A) or
RNA integrity (panel B) in MCF-7 breast tumour cells [a subtype of breast
tumour (lumina! A)]. The effect
of 0.1 AA docetaxel treatment with concurrent 10 Gray radiation is also
depicted. Radiation treatment
alone has little effect on RNA content and integrity, which was only slightly
augmented by concurrent 0.1
M docetaxel treatment.
Figure 8: Graphs of A2780 cells were treated with increasing doses of
Epirubicin with
and without 10Gy radiation. A) RIN value vs. Epirubicin Dose; B) RDI value vs.
Epirubicin Dose; C) RNA
concentration vs. Epirubicin Dose.
Figure 9: Graphs of MCF-7 cells were treated with increasing doses of
Epirubicin with
and without 10Gy radiation. A) RIN value vs. Epirubicin; B) RDI value vs.
Epirubicin; C) RNA
concentration vs. Epirubicin Dose.
Figure 10: Graphs of A2780 cells treated with 5 nM Docetaxel, 10 Gy Radiation
and 10
Gy Radiation + 5 nM Docetaxel at 72 hours and 96 hours. A) Mean RIN value with
drug and radiation
treatment (n=3). Error bars represent std deviation; B) Mean RDI value with
Docetaxel and radiation
treatment (n=3) suggest an increase in RDI value with radiation+Docetaxel
compared with Docetaxel
alone at 96 hr. Error bars represent standard deviation.
Figure 11: Graphs of RDI versus RNA concentration of pCR responders and pCR
non-
responders for samples taken (A) mid therapy and (B) post therapy.
Figure 12: is a graph of an example electropherogram in which the 18S peak,
the 28S
peak, the intermediate banding region, the low C banding region, the low B
banding region, the low A
banding region and the marker region are defined.
Figure 13: is an image of a gel of electrophoretically separated RNA samples
and
corresponding RIN and RDI values for each sample.
Detailed description of the Disclosure
I. Definitions
[00075]
The term "amount" as used herein with respect to RNA degradation refers to an
amount
(e.g. relative amount or absolute amount) of RNA degradation that is
detectable or measurable in RNA
isolated from a sample. For example, the amount can be expressed using an
absolute value (e.g. an RDI
value or a RIN value) or a relative amount such as 1.2, 1.3, 1.4, 1.5, 1.6,
1.7, 1.8, 1.9, 2.0, 2.2, 2.4, 2.6,
2.8, 3.0, 3.2, 3.4, 3.6, 3.8, 4.0, 4.2, 4.4, 4.6, 4.8, 5.0, 10, 15, 20, 25,
30, 40, 60, 80 and/or 100 times a
control amount, where for example, the control amount is, for example, a pre-
treatment amount or, for
12
7944618
Date Recue/Date Received 2022-10-28
example, a reference value corresponding to the average or median level in
untreated or pretreatment
samples. The amount can be compared to one or more threshold values, for
example which identities
subjects with a likelihood of responding to the treatment.
[00076]
The term "baseline amount" as used herein refers to an amount of RNA
degradation
(e.g. how intact the RNA is) in a sample such as a pretreatment sample that
can be used for comparison
to a test sample (e.g. comparison to a cell population and/or tumour) taken at
a later time point, for
example during or after treatment e.g. during or after a treatment regimen
comprising radiation and
optionally chemotherapy, cytotoxic antibody and/or other treatment. For
example, in methods related to
monitoring response to treatment, "base-line amount" can refer to a level of
RNA degradation in a sample
taken prior to a subsequent sample, e.g. a base-line sample is taken before
treatment, the comparison to
which provides an indication of response to treatment.
[00077]
The term "assaying a cancer cell sample to obtain a RNA integrity value" as
used herein
means performing an assay on a RNA sample of the cancer cell sample for
ascertaining or measuring
quantitatively or semi-quantitatively the degree of degradation and/or
intactness of RNA or a fraction
thereof to provide a value representative of the assay results. For example,
the RNA integrity value can
be determined by a number of methods involving microcapillary electrophoresis,
for example by
determining a RNA integrity number using for example an Agilent Bioanalyzer
machine, an Ex perion
Capillary Electrophoresis System with its equivalent RNA Quality Index (RQI),
Nanodrop (Thermo
Scientific, Inc.) or other equivalent systems, such as those manufactured by
Applied Biosystems, Lumex,
or Beckman Coulter Corporation or similar system, and/or for example In where
the RNA integrity value
is a 285:18s rRNA ratio, determined using gel agarose electrophoresis and/or
spectroscopy, for example
by assessing UV absorbance at 280:260. Other methods of determining RNA
integrity including methods
disclosed in Patent Cooperation Treaty Application No PCT/CA2013/000408
entitled ASSAYS,
METHODS AND APPARATUS FOR ASSESSING RNA DISRUPTION filed April 24, 2013,
incorporated
by reference herein in its entirety, describes assays for assessing RNA
integrity, which can be used
herein. In embodiments, an electropherogram dataset is obtained (e.g. features
of which comprise the
RNA profile) and used to calculate an RNA integrity value.
[00078]
The term "assaying a cancer cell sample to obtain a RNA concentration" as used
herein
means performing an assay on a RNA sample for ascertaining or measuring
quantitatively the amount of
RNA or a fraction thereof to provide a value representative of the assay
results. For example, the RNA
concentration can be determined by a number of methods including for example
microcapillary
electrophoresis, for example using for example an Agilent Bioanalyzer machine,
an Experion Capillary
Electrophoresis System with its equivalent RNA Quality Index (RQI), Nanodrop
(Thermo Scientific, Inc.)
or other equivalent systems, such as those manufactured by Applied Biosystems,
Lumex, or Beckman
Coulter Corporation or similar system. The RNA concentration can be based on
UV absorbance, for
example by assessing UV absorbance at 260 nm.
13
7944618
Date Recue/Date Received 2022-10-28
[00079] The term
"breast cancer" as used herein includes all subtypes of breast cancer,
including
the HER2, basal, lumina! A, luminal B, and "normal" subtypes of breast cancer
as well as locally
advanced breast cancer (LABC).
[00080]
The term "cancer cell sample" as used herein means for example any sample
comprising
cancer cells and/or cancer bed cells including for example cells obtained from
a cancer cell source such
as tumour tissue or an in vitro cell culture. The cancer cells can for example
be breast cancer cells,
ovarian cancer cells, non-melanoma skin cancer, head and neck cancer, breast
cancer, lung cancer cells
such as non-small cell lung cancer, cervical cancer, anal cancer, prostate
cancer, lymphoma cancer
cells, liposarcoma cancer cells, or any cancer cells treatable by a regimen
comprising radiation or being
tested for treatability by a regimen comprising radiation.
[00081] The term
"cancer bed cells" as used herein means reparative tissue cells that have
infiltrated a site previously occupied by a cancer (e.g. at the tumour lesion
site) and can include stromal
connective tissues surrounding the tumour. These may be obtained in a patient
who has responded to
the radiation treatment, optionally post treatment or during treatment in a
patient that has responded
rapidly. For example in the MA22 study, some tumours that exhibited a pCR post-
treatment had strong
RNA degradation in samples obtained at the tumour lesion site, even though no
tumour cells were
present, and in the MA22 study some samples that went on to have a pCR had no
tumour cellularity mid-
therapy but had a high RDI value (low RIN) which indicates that the cells
providing the RNA at this
timepoint were likely reparative tissue cells. These cancer bed cells are more
metabolically active than
the non-tumour cells which surround the site.
[00082] The term
"changing cancer treatment" as used herein includes for example one or more
of changing the dosage level and/or schedule of the radiation or
chemotherapeutic, discontinuing the
treatment, adding a chemotherapeutic agent(s), biologic(s) or radiosensitizing
agent(s) to the treatment
or changing to an alternate cancer treatment such as a drug therapy or surgery
(e.g. discontinuing
radiation treatment).
[00083] The term
"comparable to a control" or "comparable to a reference threshold" or "absence
of a change" as used herein in the context of a RNA integrity value or RNA
concentration means less
than a 30% decrease or increase, optionally, this decrease could be less than
a 25% decrease, less than
a 20% decrease, or less than a 10% decrease from a control such as a
pretreatment control, for example
, or the increase could be less than a 25% increase, less than a 20% increase,
or less than a 10%
increase from a control such as a pretreatment control. In an in vitro method,
"comparable to a control"
"comparable to a reference threshold" or "absence of a change" can be less
than a 10% or 5% decrease
or increase. In an in vivo method, "comparable to control", "comparable to a
reference threshold" or
"absence of a change" can be less than a 30% or 25% increase or decrease.
[00084]
The term "control" as used herein refers to a comparator sample such as intact
RNA
and/or RNA obtained from a subject or a group of individuals who are known as
non-responders and/or
responders and/or a reference sample such as a pretreatment sample or earlier
sample from the tested
14
7944618
Date Recue/Date Received 2022-10-28
individual. For example, the control can be a sample from a subject comprising
cancer cell RNA such as
breast cancer or ovarian cancer cell RNA, such as a pretreatment or earlier
sample from the subject. The
control can also be the expected or predetermined RNA integrity value for an
untreated cancer tissue or
cancer cell RNA sample (e.g. intact RNA). For example, an untreated cancer
tissue or untreated cancer
cell RNA sample can be determined and is expected to be "intact", for example
have a RIN value of
greater than 7, or greater than 8, or greater than 9, (a RIN of 9 can be used
particularly for in vitro
cultures). The control can be used as a reference threshold for example when
monitoring the response of
a patient or in cell line based testing. The baseline or pretreatment levels
seen in cells in in vitro cultures
is a much lower level of RNA degradation than in vivo, where tumours can have
significantly higher
baseline levels of RNA degradation. Cell cultures in the laboratory are under
ideal conditions to promote
cell viability, including growth factors, nutrients, buffers, appropriate
oxygenation, ROS scavengers,
vitamins, etc. In an embodiment, the control is a positive control e.g.
cells/tumour treated with radiation.
In another embodiment, the control is a negative control, e.g. cell/tumour not
receiving radiation
treatment, optionally not receiving any treatment.
[00085]
The term "cytotoxic treatment" as used herein means any agent that can be used
with
radiation, sequentially and/or concurrently, that can induce cell death that
is used in the treatment of
cancer, including for example, traditional and non-traditional chemotherapy
(e.g. targeted therapies),
radiation treatment, hormonal treatment (e.g. for responsive cancers) and
combinations thereof. Such
agents include but are not limited to microtubule stabilizing agents such as
Docetaxel and paclitaxel,
DNA synthesis inhibitors such Epirubicin, inhibitors of Her2 Receptor such as
Trastuzumab, DNA cross-
linking agents such as Mafosfamide, carboplatin and cisplatin, VEGFA
inhibitors such as Bevacizumab,
Receptor Tyrosine Kinase inhibitors such as Sunitinib and Toceranib,
Bisphosphonates such as
Zoledronic acid, Thymidylate synthase inhibitors such as 5-fluorouracil.
[00086]
The term "chemotherapeutic agent" as used herein means any drug or drug
combination
used for the treatment of cancer such as breast cancer or ovarian cancer or
other cancers treatable by
radiation comprising regimens such as for example non-melanoma skin cancer,
head and neck cancer,
breast cancer, non-small cell lung cancer, cervical cancer, anal cancer,
prostate cancer, including for
example drugs used for primary chemotherapy including for example a
platinating agent (e.g. cisplatin
and/or carboplatin) and/or a taxane (e.g. paclitaxel and/or docetaxel), drugs
typically used in the
treatment of recurrent ovarian cancer (e.g. anthracyclines such as doxorubicin
or epirubicin or their
pegylated forms), topoisomerase I and ll inhibitors (e.g. topotecan and
etoposide, respectively),
nucleoside analogs such as gemcitabine and 5-fluorouracil, DNA cross-linking
agents such as
Mafosfamide, the estrogen receptor blocker tamoxifen, and/or the Her-2/Neu
blocker bevacizumab. In an
embodiment, the chemotherapeutic agent is sunitinib.
[00087]
The term "dose" as used herein in reference to radiation refers to an
individual radiation
exposure either administered at each time within a schedule or the total
amount of radiation exposure
within a schedule. With respect to a chemotherapy treatment a dose means an
amount of an individual
7944618
Date Recue/Date Received 2022-10-28
drug either administered at each time within a schedule OR the total amount of
each drug administered
within a schedule or the total amount of drug administered during a course of
chemotherapy.
[00088]
The term "decreased RNA integrity" as used herein for determining response
with
respect to patient samples means an RNA integrity that is at least 20%, at
least 25%, at least 30%, at
least 40%, at least 50%, at least 60%, at least 70%, at least 80% or at least
90% less than a control or
corresponding value for example a pretreatment sample or for example a maximal
value (e.g. maximal
RIN). With respect to a cell culture sample, a decrease of at least 5%, 10%,
15%, 20% or 25% compared
to control is indicative of a decreased RNA integrity.
[00089]
The term "decreased RNA concentration" as used herein means an RNA integrity
that is
at least 20%, at least 25%, at least 30%, at least 40%, at least 50%, at least
60%, at least 70%, at least
80% or at least 90% less than a control or corresponding reference value for
example a pretreatment
sample or for example a maximal value.
[00090]
The term "positive treatment outcome" in the context of a patient as used
herein refers
to a positive therapeutic response to treatment, for example alleviation or
amelioration of one or more
symptoms or conditions, diminishment of extent of disease, stabilized (i.e.
not worsening) state of
disease, preventing spread of disease or preventing disease progression, delay
or slowing of disease
progression, reversal of disease, amelioration or palliation of the disease
state, and remission (whether
partial or total), whether detectable or undetectable. "A positive treatment
outcome" can also mean
prolonging overall survival, stable disease and/or disease/progression free
survival as compared to
expected survival if not receiving treatment, including for example a
pathologic complete response post-
treatment. The extent of the positive treatment outcome can be for example
related to the extent of RNA
degradation and/or RNA concentration decrease determined for a cancer sample
obtained during
treatment.
[00091]
The term "post-treatment" as used herein means after completion of a treatment
comprising a radiation regimen, optionally completion of all arms of a
treatment regimen e.g. after
completion of any chemotherapy or other treatment post radiation.
[00092]
The term "negative treatment outcome" in the context of a patient as used
herein means
a lack of a therapeutic response to the treatment, for example no response,
recurrence of disease, or
spread of disease (disease progression).
[00093]
The term "responders" as used here means patients that demonstrate a positive
treatment or therapeutic outcome, including for example, a measurable
therapeutic response.
Responders optionally include patients who demonstrate increased disease free
survival (DFS) or overall
survival (OS) compared to the average for a group of similarly treated
patients, a pathological complete
response (pCR) or a partial response.
[00094]
The term "non-responders" as used herein means patients (e.g. non-responders)
that do
not demonstrate a positive treatment outcome including for example no
measurable therapeutic
16
7944618
Date Recue/Date Received 2022-10-28
response, for example exhibit a negative therapeutic outcome such as a
decreased DSF or OS
compared to the average for a group of similarly treated patients.
[00095] The term "RNA sample" means any sample comprising purified
and/or isolated RNA,
including any purified and/or isolated RNA fraction such as total RNA, rRNA,
and/or mRNA. In an
embodiment, the RNA sample comprises rRNA. An RNA sample can be obtained for
example using for
example a number of methods known in the art for isolating RNA.
[00096] The term "ovarian cancer" as used herein means all subtypes of
ovarian cancer,
including the serous, clear cell, endometrioid, and mucinous subtypes [10].
[00097] The term "radiation" in relation to a treatment means any
energy, photon or particle,
applied to a tumour, including for example ionizing radiation.
[00098] The term "radiosensitizer" as used herein means an agent such as a
small molecule
drug or biologic that makes a tumour cell more sensitive to radiation therapy.
The radiosensitiver can be
an oxygen increasing agent, a hypoxic cell sensitizer, a halogenated
pyrimidine, or a bioreductive agent.
[00099] The term "oxygen increasing agent" as used herein means an
agent such as a small
molecule drug or biologic that increases tumour oxygenation and/or reduces
tumour hypoxia, including
for example oxygen. The term "hypoxic cell sensitizer" as used herein means an
agent, such as a small
molecule drug, that can mimic oxygen (but does not increase oxygen levels) and
that is typically not
metabolized by tumour cells allowing the agent to penetrate a tumour. Examples
include nitroimidazoles.
Hypoxic cell sensitizers, such as 2-nitroimiazole increase the killing of
hypoxic cells, which are typically
rad ioresistant.
[000100] The term "bioreductive agent" as used herein means an agent such as a
small molecule
drug that can be reduced to a cytotoxic species, for example in a hypoxic
region of a tumour. Examples
include quinone antiobiotics such as mitomycin C, niroaromatics, and di-N
oxides.
[000101] The term "radioprotector" as used herein means s used herein means an
agent such as
a small molecule drug or biologic that makes a tumour cell less sensitive to
radiation therapy.
[000102] The term "RNA degradation" as used herein means a decrease in the RNA
integrity of
isolated cancer cell or tissue RNA compared to RNA from untreated cancer cells
or tissues, or an
expected RNA integrity for cancer cells. For example, human RNA (e.g. isolated
from primary cells
and/or tissue) is commonly recognized as degraded when RIN is <7, and/or
optionally <7, less than 6.8,
less than 6.6, less than 6.4, less than 6.2, less than, 6.0, less than 5.8,
less than 5.6, less than 5.4, less
than 5.2 or less than 5.0 and for cell lines when RIN is for example =<8,
optionally <9. The RNA
degradation can comprise treatment induced RNA degradation and autolytic RNA
degradation.
[000103] The term "autolytic RNA degradation" as used herein refers to RNA
degradation taking
place during autolytic cell destruction. Autolysis is initiated, for example,
by the cells' lysosomes releasing
digestive enzymes into the cytoplasm due to the cessation of active processes
in the cell, and not due to
17
7944618
Date Recue/Date Received 2022-10-28
an active physiologic or pathophysiologic process. Autolytic RNA degradation
in a sample can be
induced by removal of cells from physiologic environment for extended period
of time (e.g. incubation in
saline) and/or nonspecific cessation of physiologic processes e.g. heat
treatment.
[000104] The term "treatment induced RNA degradation" as used herein refers to
discretely
fragmented and/or degraded RNA that is signal induced in response to a
cytotoxic treatment such as
radiation treatment, chemotherapy treatment and/or cytotoxic antibody
treatment (e.g. Trastuzumab).
Cytotoxic signal induced RNA degradation can include RNA degradation that has
some features that
resemble autolytic degradation, particularly for example during later stages.
[000105] The term "RNA integrity" as used herein means to the degree of
intactness of the RNA
following extraction or isolation from the cell or tissue sample e.g. whether
the isolated RNA is degraded.
RNA integrity can be assigned a value (e.g. RNA integrity value) that
corresponds to the degree of RNA
intactness and/or degradation, e.g. proportionately or inverse
proportionately. A high RNA integrity is
commonly taken as meaning little or no degradation, for example less than a
30% decrease, less than
25% decrease, less than 20% decrease from maximal RNA integrity value (e.g.
when the RNA integrity
value is measured using RIN, e.g. RIN=10 or other scale where RNA integrity
value is proportional to the
RNA integrity of the sample), or less than a 30% increase, less than a 20%
increase for example from
baseline when the RNA integrity value is inverse proportional to the RNA
integrity of the sample e.g.
RDI (e.g. for an in vivo sample and optionally less than 10% or 5% decrease or
increase for an in vitro
sample) and/or a RNA integrity value corresponding to a control such as a
pretreatment control and
retention of capacity to amplify mRNAs of interest following extraction or
isolation. Using RDI, a high RNA
integrity is for example less than 10, or less 3 and using RIN a high RNA
integrity is greater than 7,
greater than 8 or greater than 9. A low RNA integrity is for example RNA that
exhibits greater than 20%,
greater than 25%, greater than 30%, greater than 35%, greater than 40%,
greater than 45%, greater
than 50%, greater than 55%, greater than 60%, greater than 65%, greater than
70%, or greater than
75% decrease from maximal for example using RIN, wherein maximal RIN=10 (or
other scale where the
RNA integrity value is proportional to RNA integrity), or exhibits greater
than 20%, greater than 25%,
greater than 30%, greater than 35%, greater than 40%, greater than 45%,
greater than 50%, greater
than 55%, greater than 60%, greater than 65%, greater than 70%, or greater
than 75% increase in
RNA integrity value for example from baseline, for example using RDI (or other
scale where the RNA
integrity value is inverse-proportional to RNA integrity), a control such as a
pretreatment control or
decreased capacity to amplify mRNAs of interest when they are known to be
present in controls in RNA
following extraction or isolation.
[000106] The term "RNA integrity value" as used herein is a measure of the
degree or extent of
intactness or degradation of RNA of a sample following extraction or isolation
from the cell or tissue
sample. For example the RNA integrity value can be a spectrophotometer
intensity measurement, and/or
a 286:18S ribosomal RNA ratio. The RNA integrity value can be an RNA integrity
number (RIN),
determined using an Agilent Bioanalyzer, the output of the RIN algorithm,
wherein generally the higher
the RIN value, the higher the RNA integrity and the lower the RIN value the
higher the RNA degradation.
18
7944618
Date Recue/Date Received 2022-10-28
For example, a RIN of 0 represents completely hydrolyzed RNA and a RIN of 10
represents completely
undegraded RNA. A person skilled in the art would recognize that the scale
could also be inverted (for
example, by dividing the value into 1, e.g. 1/RNA integrity value) such that
the lower the RNA integrity
value, the higher the RNA integrity and the higher the RNA integrity value,
the lower the RNA integrity.
For example, the RNA integrity value can be the output of a calculation based
on electropherogram
features as described in Patent Cooperation Treaty Application No
PCT/CA2013/000408, ASSAYS,
METHODS AND APPARATUS FOR ASSESSING RNA DISRUPTION filed April 24, 2013, for
example
represented as an RNA Disruption Index (RDI), or can be a transformed scale.
For example for patient
samples, the scale can be defined by clinical RNA disruption assay (RDA) zones
based on cut-offs or
response thresholds, wherein each score or RDA zone is associated with a
likelihood of a response for
example associated with a likelihood of responsive to a treatment, for example
defined by NPV and/or
PPV. In embodiments wherein RDI or RDA zones are employed, a high or increased
RNA integrity value
(for example compared to a baseline sample or control) is indicative of high
or increased RNA
degradation. For example in such an embodiment, the higher the RNA integrity
value, the greater the
induced RNA degradation (and decrease in RNA integrity). Any scale can be
employed, for example 3,
10, or 600. Accordingly, wherein the RNA integrity value is proportional to
the RNA integrity of the
sample, a high or increased RNA integrity value corresponds to a high or
increased RNA integrity and
wherein the RNA integrity value is inverse proportional to the RNA integrity
of the sample, a high or
increased RNA integrity value corresponds to low or decreased RNA integrity.
The RNA integrity value
can be calculated using one or a number of features. Also the scale employed
can be a linear scale or a
logarithmic scale.
[000107] The term "RNA isolating or stabilizing composition" as used herein
refers to any
composition that inhibits RNAse activity and/or stabilizes RNA preventing RNA
degradation.
[000108] The term "stable RNA integrity" as used herein means RNA that is not
degraded
appreciably, for example as compared to an appropriate comparator sample or
the expected RNA
integrity for the cell type of tissue. Typically for humans this is isolated
RNA with RIN=>7, and can be for
example in the context of tumour cell RNA ->6.8, -------------------------- -
>6.6, ->6.4, ->6.2, ->6.0, or =>5.8 and for cell lines,
for example RIN=>9. Other measures including RDI can also be used.
[000109] The terms "patient" and "subject" which are used herein
interchangeably refer to any
member of the animal kingdom, including mammals, for example dogs, rodents
such as rats and mice,
preferably a human being, including for example a subject that has or is
suspected of having cancer,
optionally breast cancer or ovarian cancer.
[000110] The term "RNA" as used herein includes any RNA or RNA fraction,
including but not
limited to total RNA, rRNA and/or mRNA, of subset of RNAs for example RNA can
include the total of
RNA types and components that may be present following RNA isolation, for
example ribosomal RNAs
(rRNAs) messenger RNAs (mRNAs), or fractions comprising for example at least
rRNA. As an example,
total RNA can be used with the methods described herein or a class or subset
of RNAs can also be used.
19
7944618
Date Recue/Date Received 2022-10-28
For example, RNA subsets that can be assayed with the methods described
include for example subsets
comprising rRNA, and/or mRNA.
[000111] The term "resistant" as used herein refers to a cancer cell or tumour
response to
radiation alone or in combination with a cytotoxic, optionally
chemotherapeutic, agent or regimen, where
the cancer cells or subset of cancer cells within a tumour show no or
insufficient response (for example
determinable in a clinical study) to the treatment in terms of RNA
degradation, which is associated for
example with a negative treatment outcome.
[000112] The term "response" as used herein refers to a cancer cell or tumor
response to
radiation alone or in combination with a cytotoxic, optionally
chemotherapeutic, agent and/or regimen,
where the cancer cells or subset of cancer cells within a tumour respond to
the treatment in terms of RNA
degradation ¨ e.g. show significant and/or sufficient RNA degradation (for
example determinable in a
clinical study), which is associated for example with a positive treatment
outcome.
[000113] The term "low risk" as used in relation to progression refers to less
than average risk
(e.g. decreased probability) calculated for a group of patients with the same
cancer, treated similarly; and
high risk of progression means greater than average risk (e.g. increased
probability) compared to the
group of patients.
[000114] The term "RNA Disruption Index" or "RDI" is used herein as used in
Patent Cooperation
Treaty Application No PCT/CA2013/000408 filed April 24, 2013 and is a value
generated using RNA
disruption assay (RDA) and can be a ratio of defined features (e.g. features
shown in figure 12), of the
output of linear discriminant analysis (LDA) or quadratic discriminant
analysis of features described
herein. The RDI is a logarithmic scale and can be calculated using features
shown for example in Figure
12, optionally for example based on (Intermediate Area+LowC Area)/(28S
Area+18S Area). The RDI
scale reflects the absolute ratio of disrupted RNA/normal RNA. The RDI values
determined from a group
of known response outcome patients (e.g. that are determined to be associated
with a particular NPV or
PPV) can be used to define the thresholds boundary for RDA zones.
[000115] The term "RDA zones" as used herein refers to clinical zones
associated with treatment
response outcome comprising a range of RDI values. Each RDA zone is defined by
one or two
boundaries each boundary corresponding to a selected threshold (e.g.
corresponding to a desired NPV
or PPV). Subject RDI values that fall within the clinical zones that are
associated with or are predictive of
treatment response, for example pCR and/or increased DFS. In an embodiment, 3
RNA disruption assay
zones can be used, RDA zone 1, RDA zone 2 and RDA zone 3, defined by selected
NPV and/ or PPVs.
A person skilled in the art would readily realize that any number of zones can
be used each with different
selected thresholds.
[000116] The term "RDA zone 1" as used herein refers to a range of RDA scores
(e.g. RDI values)
that have a negative predictive value (NPV) of at least 0.8, at least 0.85, at
least 0.9, at least 0.95, at
least 0.96, at least 0.97, at least 0.98 or greater. These numbers and the
associated thresholds, are for
example based on DES and/or pCR clinical trial results for a specific
treatment for example such as a
7944618
Date Recue/Date Received 2022-10-28
clinical trial such as a clinical trial described in Example 1 with increased
number of patients. In an
example embodiment, RDA Zone 1 is equal to an RDI of equal or less than 10
calculated using the
feature Intermediate Area/(28S+18S Areas). In other embodiments, other
features or combination of
features can be used. The "RDA zone 1" can be defined to include any set of
scores by selecting the
desired NPV.
[000117] The term "RDA zone 2" as used herein refers to a range of RDA scores
(e.g. RDI values)
falling between RDA zone 1 and 3, and can be considered an intermediate or
indeterminate zone.
[000118] The term "RDA zone 3" as used herein refers to a range of RDA scores
(e.g. RDI values)
that have a positive predictive value (PPV) of at least 0.15, at least 0.16,
at least 0.17, at least 0.18, at
least 0.19, at least 0.2 or greater. These numbers and the associated
thresholds are for example based
on pCR and/or DFS clinical trial results for a given treatment, e.g. a
clinical trial as described in Example
1 with additional patients. The "RDA zone 3" can be defined to include any set
of scores by selecting the
desired PPV.
[000119] In an embodiment, the radiation response score is an RDA zone 1, 2 or
3. The radiation
response score can also be resistant, responding and indeterminate.
[000120] The term "response threshold" or "reference threshold" as used herein
can be an
expression or cut-off value of RNA integrity and/or RNA concentration in a pre-
determined sample or
group of samples, above and/or below which a cancer cell type or tumour is
identified as being more
likely resistant or more likely responsive to treatment and for example
indicative of patient outcome. For
example, a patient that has a RNA integrity value indicative of RNA integrity
and/or RNA concentration
below a cut-off or response threshold is indicated to be more likely
responsive to the radiation treatment
and/or regimen and/or predicts positive treatment outcome. The response
threshold can for example be
derived from a control such as a pretreatment or untreated sample or a value
derived from a population
of subjects that are known responders and/or non-responders which for a
preselected degree of
specificity and sensitivity, classifies patients likely to be responders from
patients likely to be non-
responders. In the case of a control such as a pretreatment control or
untreated control, the response
threshold can be the expected RNA integrity for (e.g. when a RIN value,
optionally 7, 8, 9 or 10) based on
for example the cell population, treatment etc. A person skilled in the art
will recognize that the direction
of the change indicative of worse or better outcome will depend on the scale
used.
[000121] The term "sample" as used herein refers to 1) any biological fluid,
cell or tissue sample
from a subject (e.g. test subject) or cell line that comprises cellular RNA,
optionally tumour tissue/cells
and/or cancer bed cells and/or 2) RNA derived from such a sample. For example,
the sample can be a
biopsy, including a needle aspirate, such as a fine needle aspirate, a core
biopsy, a brush biopsy and/or
a laparoscopic biopsy. The sample can, for example, be a "post-treatment"
sample wherein the sample is
obtained after one or more doses of radiation or cytotoxic cancer treatments,
or a "base-line sample"
which is optionally pre-treatment or taken at an earlier time-point than the
post-treatment sample, and is
21
7944618
Date Recue/Date Received 2022-10-28
for example, used as a base line for assessing or monitoring response to a
radiation optionally in
combination with cytotoxic treatment.
[000122] As used herein, and as well understood in the art, "treatment" is an
approach for
obtaining beneficial or desired results, including clinical results.
Beneficial or desired clinical results can
include, but are not limited to, alleviation or amelioration of one or more
symptoms or conditions,
diminishment of extent of disease, stabilized (i.e. not worsening) state of
disease, preventing spread of
disease, delay or slowing of disease progression, reversal of disease,
amelioration or palliation of the
disease state, and remission (whether partial or total), whether detectable or
undetectable. "Treatment"
can also mean prolonging survival as compared to expected survival if not
receiving treatment.
[000123] In understanding the scope of the present disclosure, the term
"comprising" and its
derivatives, as used herein, are intended to be open ended terms that specify
the presence of the stated
features, elements, components, groups, integers, and/or steps, but do not
exclude the presence of other
unstated features, elements, components, groups, integers and/or steps. The
foregoing also applies to
words having similar meanings such as the terms, "including", "having" and
their derivatives. Finally,
terms of degree such as "substantially", "about" and "approximately" as used
herein mean a reasonable
amount of deviation of the modified term such that the end result is not
significantly changed. These
terms of degree should be construed as including a deviation of at least 5%
of the modified term if this
deviation would not negate the meaning of the word it modifies.
[000124] The recitation of numerical ranges by endpoints herein includes all
numbers and
fractions subsumed within that range (e.g. Ito 5 includes 1, 1.5, 2,2.75, 3,
3.90,4, and 5). It is also to be
understood that all numbers and fractions thereof are presumed to be modified
by the term "about."
Further, it is to be understood that "a," "an," and "the" include plural
referents unless the content clearly
dictates otherwise.
[000125] Further, the definitions and embodiments described in particular
sections are intended to
be applicable to other embodiments herein described for which they are
suitable as would be understood
by a person skilled in the art. For example, in the following passages,
different aspects of the invention
are defined in more detail. Each aspect so defined may be combined with any
other aspect or aspects
unless clearly indicated to the contrary. In particular, any feature indicated
as being preferred or
advantageous may be combined with any other feature or features indicated as
being preferred or
advantageous.
II. Methods
[000126] Described herein are methods for evaluation of cancer cells including
for example breast
cancer cells and ovarian cancer cells for their response to radiation
treatment.
[000127] A clinical study was described in PCT/CA2008/001561, herein
incorporated by
reference, which demonstrated that breast cancer cells demonstrate reduced RNA
integrity when treated
with a chemotherapeutic. It is has also been demonstrated by the inventors
that ovarian cancer cell lines
22
7944618
Date Recue/Date Received 2022-10-28
show loss of RNA content (e.g. concentration) and/or integrity in response to
chemotherapeutic
treatment.
[000128] Patent Cooperation Treaty Application No PCT/CA2013/000408 entitled
ASSAYS,
METHODS AND APPARATUS FOR ASSESSING RNA DISRUPTION filed April 24, 2013,
incorporated
by reference herein in its entirety, describes assays for assessing RNA
integrity, which can be used
herein.
[000129] Accordingly, an aspect includes a method of evaluating a cancer cell
sample optionally
selected from a breast cancer cell sample and an ovarian cancer cell sample,
the method comprising:
a) obtaining a cancer cell sample, optionally a breast cancer cell sample or
an ovarian
cancer cell sample, after the cancer cells have been exposed to a radiation
dose;
b) assaying the cancer cell sample to obtain a RNA integrity value and/or a
RNA
concentration of the cancer cell sample.
[000130] A decreased RNA integrity value is reflective of RNA degradation and
responsiveness to
treatment.
[000131] In an embodiment, the method further comprises comparing the RNA
integrity value
and/or the RNA concentration of the cancer cell sample to a response threshold
wherein a decreased
RNA integrity value and/or RNA concentration of the cancer cell sample
compared to the response
threshold is predictive that the cancer cells are responsive to the radiation
therapy and a comparable or
an increased RNA integrity value (i.e. indicative of comparable or increased
RNA integrity) and/or RNA
concentration of the cancer cell sample compared to the response threshold is
predictive the cancer cells
are unresponsive to the radiation treatment.
[000132] It is demonstrated herein that RNA degradation can be detected in
breast cancer cells
and ovarian cancer cells in response to radiation exposure. In an embodiment,
the cancer cell sample is
a breast cancer cell sample. In another embodiment, the cancer cell sample is
an ovarian cancer cell
sample.
[000133] RNA degradation was detected for example after one dose in the
ovarian cancer cell
line. In an embodiment, the cancer cells have received 1, 2, 3, 4, 5, 6 or
more doses of radiation
treatment.
[000134] RNA degradation is detected in cancer cells such as breast cancer and
ovarian cancer
cells treated with radiation and a cytotoxic agent such as a chemotherapeutic.
In an embodiment, the
cells are further treated with a dose of a cancer cytoxic agent, optionally a
chemotherapeutic agent. In an
embodiment, the chemotherapeutic agent is selected from anthracyclines,
taxanes and combinations
thereof, preferably wherein the chemotherapeutic agent comprises epirubicin,
docetaxel or combinations
thereof. In another embodiment, the taxane is paclitaxel, docetaxel,
larotaxel, AbraxaneTM,
23
7944618
Date Recue/Date Received 2022-10-28
docoxahexaenoic acid-linked paclitaxel, paclitaxel polyglumex, Ortataxel,
GenexolTM, liposomal-
encapsulated paclitaxel, and paclitaxel in a Vitamin E emulsion In a further
embodiment, the taxane is
docetaxel or paclitaxel.
[000135] In an embodiment, the patient is administered in a radiosensitizer
agent. In an
embodiment, the radiosensitizer agent is selected from a variety of classes of
compounds, including
oxygen, hypoxic cell sensitizers, halogenated pyrimidines, and bioreductive
agents.
[000136] A decreased RNA integrity value is reflective of RNA degradation,
primarily the
degradation of the highly abundant rRNAs. In an embodiment, the RNA sample is
total RNA. In another
embodiment, the RNA sample is and/or comprises rRNA. In another embodiment,
the RNA sample is
and/or comprises mRNA.
[000137] The method can be used for example to test a tumour's sensitivity to
a radiation
sensitizing agent and/or screen for compounds that radio-sensitizes cancer
cells or otherwise improve
response to radiation treatment. For example, cells can be pretreated in vitro
with a compound and
subsequently exposed to radiation. The extent of RNA integrity and
concentration changes can be used
to screen compounds. Accordingly, in another embodiment, the cells are
contacted with a compound to
determine if the compound is a radio-sensitizer and/or a radio-protector. As
demonstrated in the
examples, combinations of chemotherapy and radiation can result in prominent
increases in RNA
degradation compared to chemotherapy treatment or radiation treatment alone.
Accordingly in an
embodiment, the method is for identifying synergistic combinations of a test
agent and radiation.
[000138] In an embodiment, the method comprises: i) exposing cells to a fixed
dose or radiation
and an increasing dose of the test agent and ii) assessing the RNA integrity
of the cells, wherein a test
compound that reduces the RNA integrity compared to a control is radiation
sensitizer and the RNA
integrity is a radiation radioprotector.
[000139] One example of a screen would be to conduct the RNA integrity assay
with fixed doses
of radiation and to assess the ability of the radioprotector or the
radiosensitizer to reduce or augment the
degree of RNA degradation, respectively. The extent of RNA degradation at the
various radiation doses
in the absence or presence of the radiosensitizer or radioprotector can be
used to determine LD5Os in
the absence or presence of the agent, in order to quantify the degree of
protection/sensitization.
[000140] Extent of response can be assessed for example by comparing to a
response threshold.
For example, the response threshold for in vitro embodiments can comprise an
untreated control or a
value that corresponds to an in vitro control. For example, cell lines
generally have little if any RNA
degradation if handled appropriately using methods which are known in the art.
The response threshold
therefore can be a selected value for example, 9, 8 or 7. In an embodiment,
the response threshold is
derived from an untreated sample and/or is a standard derived from a plurality
untreated samples.
[000141] The response threshold can also be a preselected threshold derived
for example from
treated or untreated samples, such as a median or average. In yet another
embodiment, the response
24
7944618
Date Recue/Date Received 2022-10-28
threshold is derived from an unresponsive sample and/or a responsive sample
and/or is a standard
derived from a plurality of unresponsive samples and/or responsive samples.
[000142] As mentioned above, cancer cells such as breast cancer and ovarian
cancer cells can be
screened in vitro. Tests can also be conducted in vivo using for example an
animal model where cancer
cells are implanted, the animal is administered radiation alone or in
combination with a test agent and the
response monitored. Accordingly, in another embodiment, the cancer cells are
exposed to radiation in
vitro or in vivo.
[000143] The cancer cell is in an embodiment in vivo in a patient.
[000144] In an embodiment, the patient is treated with radiation and/or
chemotherapy prior to
surgery. When administered after surgery, methods dexcribed herein can be used
to monitor response to
chemotherapy/radiation therapy for example in any recurrent tumours.
[000145] In vivo cells may experience increased signal induced RNA
degradation. Cells in culture
for example are not subjected to the anti-vascular effects of radiation
(Fenton et al., 2001; El Kaffas et al.,
2013), which could be expected to reduce RNA integrity.
[000146] As demonstrated in for example Figure 4b, patients who exhibited a
pathological
complete response (pCR) had decreased RNA integrity and decreased RNA
concentration post-
treatment when treated with FEC-D and radiation
[000147] A further aspect includes a method for evaluating a patient derived
cancer cell sample,
the method comprising:
a) obtaining a cancer cell sample from a subject after the subject has been
exposed to
a radiation dose;
b) assaying the cancer cell sample to obtain a RNA integrity value and
optionally a
RNA concentration of the cancer cell sample.
[000148] In an embodiment, the method further comprises comparing the RNA
integrity value and
optionally the RNA concentration of the patient cancer cell sample to a
response threshold wherein a
decreased RNA integrity and/or RNA concentration of the patient cancer cell
sample compared to the
response threshold is predictive that the patient is responsive to the
radiation therapy and a comparable
or an increased RNA integrity and optionally the RNA concentration of the
patient cancer cell sample
compared to the response threshold is predictive the patient is unresponsive
to the radiation treatment.
[000149] In an embodiment, the cancer cell sample is selected from a breast
cancer cell sample
or an ovarian cancer cell sample.
[000150] The cancer cell sample can be obtained from a cell line and/or
obtained from a patient.
For example, the cancer cell sample can be obtained in a cytological or
histological biopsy. The biopsy
can be for example a needle core biopsy or fine needle aspirate or a biopsy or
resection obtained during
surgery.
7944618
Date Recue/Date Received 2022-10-28
[000151] For example, for ovarian cancer cell sample can comprise peritoneal
fluid, and/or be a
cell scrape, a tumour fine needle aspirate (F NA) and/or core biopsy.
[000152] In an embodiment, the response is a therapeutic response e.g. the
breast cancer or
ovarian cancer cell response to the radiation treatment and/or regimen is
sufficient to provide a
therapeutic benefit to the subject. Therapeutic response is for example
predictive of clinical outcome post
treatment.
[000153] In an embodiment the clinical outcome is a positive treatment
outcome. In another
embodiment the treatment outcome is a negative treatment outcome.
[000154] The extent or degree of RNA degradation can be associated with
treatment response.
Extensive degradation for example can be associated with pathological complete
response. A lesser
degree of degradation can be associated with lesser than pathological complete
response. For example,
a subject may not achieve pathological complete but may exhibit some RNA
degradation during and/or
post- treatment. Such a subject may for example, be treated in a subsequent
treatment regimen less
aggressively than a subject with no response.
[000155] In an embodiment, the positive treatment outcome predicted is
complete pathologic
response following treatment. In an embodiment, the positive treatment outcome
predicted is reduced
risk of disease progression, increased likelihood of disease free survival
and/or increased overall
survival. In an embodiment, the negative treatment outcome predicted is an
increased risk of disease
progression, decreased survival and/or recurrence. The risk of disease
progression, length of disease
free survival and/or other outcomes are, for example, relative to the average
or median risk of
progression of responders and/or non-responders of patients with the same
disease and/or treatment.
[000156] As demonstrated for example in Figure 4b, non-PCR responders could be
segregated
from pathological complete responders in terms of RNA integrity alone or in
combination with RNA
concentration.
[000157] Another aspect includes a method of identifying non-responding
patients comprising:
a) obtaining a cancer cell sample from the patient after the patient has been
exposed to a
radiation dose;
b) assaying the cancer cell sample to obtain a RNA integrity value and/or a
RNA concentration;
C) comparing to a response threshold ; and
c) identifying patients having a RNA integrity value indicative of RNA
integrity and/or RNA
concentration that is above the response threshold.
[000158] In an embodiment, the cancer is selected from a breast cancer cell
sample or an ovarian
cancer cell sample.
26
7944618
Date Recue/Date Received 2022-10-28
[000159] Yet a further aspect includes, a method of predicting a treatment
outcome of a patient
having cancer such as breast cancer or ovarian cancer, the method comprising:
assaying a cancer cell
sample obtained from the subject to obtain a RNA integrity value and/or RNA
concentration for the
cancer cell sample, wherein the subject has been treated with a radiation
dose, wherein a RNA integrity
indicated by the RNA integrity value that is below a response threshold
predicts subject/cancer response
to the radiation treatment and a decreased risk of progression; and a RNA
integrity indicated by the RNA
integrity value and/or RNA concentration that is higher than the response
threshold predicts
subject/cancer resistance to the radiation treatment and an increased risk of
disease progression.
[000160] In an embodiment, a RDI value of 3 or less indicates non-response to
radiation, for
example wherein RDI is calculated based on (Intermediate Area+LowC Area)/(28S
Area+18S Area).
[000161] The methods can be employed for example in a method of treatment.
[000162] Accordingly, a further aspect includes a treatment method comprising:
a) exposing a patient with cancer, optioanally breast cancer or ovarian
cancer, to a radiation
dose;
b) obtaining a cancer cell sample after administration of to the radiation
dose,
c) assaying the cancer cell sample to obtain a RNA integrity value and/or a
RNA concentration of
the cancer cell sample; and
d) continuing the treatment when the RNA integrity (e.g. as indicated by the
RNA integrity value)
and/or the RNA concentration is decreased compared to a response threshold and
changing the
treatment when the RNA integrity value and/or the RNA concentration is
comparable or increased
compared to the response threshold.
[000163] The treatment can be changed for example by employing
radiosensitizing agents during
radiation therapy, modifying the dose and/or schedule of the radiation and/or
chemotherapy and/or
adding or changing the chemotherapy.
[000164] In an embodiment, the cancer cell sample is a breast cancer cell
sample obtained from a
breast cancer patient. In another embodiment, the breast cancer is Her2+,
basal subtype or lumina! B
subtype. In another embodiment, the breast cancer patient has locally advanced
breast cancer (LABC).
[000165] In another embodiment, the cancer cell sample is an ovarian cancer
cell sample
obtained from an ovarian cancer patient.
[000166] In another embodiment, the cancer cells are cancer selected from
endometrial cancer,
non-melanoma skin cancer, head and neck cancer, breast cancer, lung cancer
such as non-small cell
lung cancer, cervical cancer, anal cancer or prostate cancer. Radiation
sensitivity can vary, depending
upon tumour subtype for example as in lung cancer (Johung et al. 2013 Clin
Cancer Res. 19(22)) and
27
7944618
Date Recue/Date Received 2022-10-28
endometrial cancers (Ruyck et al. Int. J. Radiation Oncol.Biol Phys. 65: 1240)
as well as breast cancer
(Djuzenova et al, Radiation Oncology 8:98).
[000167] Radiation can be used for radiation sensitive tumours including for
example lymphoma,
liposarcoma, often in conjunction with chemotherapy. Radiation is often used
for localized cancers not
accessible to surgery and for local palliation of less radiosensitive cancers
that have spread and are
resistant to chemotherapy. Accordingly in an embodiment, the cancer cells are
cancer selected
lymphoma and liposarcoma.
[000168] RNA integrity may be used to predict response in patients. Breast
cancer and ovarian
cancer cells have increased RNA degradation even in the absence of drug; seen
for example in MCF-7
and A2780 cells described in the Examples.
[000169] MCF-7 and SkBr3 cells both show signal induced RNA degradation
effects which are
detected by RDI. MCF-7 cells also have comparable RNA degradation effects
caused by Epirubicin
and/or Docetaxel.
[000170] Breast cancer tumours have variable responses to radiotherapy63.
Molecular
signatures/heat shock protein expression are currently under investigation as
indicators of radiation
response. Markers that can identify which tumours are responding would be
useful.
[000171] Radiation can cause DNA damage and RNA damage 64'65.
[000172] Radiation induced RNA damage has been identified in lymphocytes, MCF-
7 cells66,
and HeLa cells67.
[000173] In an embodiment, the response threshold comprises a RNA integrity
value. In a further
embodiment, the response threshold comprises both a RNA integrity value and a
concentration value.
[000174] The response threshold can be derived from an untreated cancer
patient such as a
breast cancer or ovarian cancer patient or be a standard derived from a
plurality of untreated cancer
patients (e.g. cells derived from untreated breast cancer or ovarian cancer
patients or obtained prior to
treatment e.g. from one or more pretreatment samples e.g. controls).
[000175] For example, the response threshold corresponds to a pretreatment RNA
integrity value
and optionally RNA concentration. For example, the response threshold can
correspond to the RNA
integrity value of a biopsy RNA sample taken from the subject or two or more
subjects prior to initiating
therapy or reference values from tumours of similar patients prior to therapy.
A decrease in RNA integrity
compared to the pretreatment RNA integrity value for example, would indicate
treatment response and/or
positive treatment outcome and for example a comparable RNA integrity value of
the cancer cell RNA to
the pretreatment RNA integrity value, would indicate resistance negative
treatment outcome.
[000176] The response threshold can also be derived a patient whose response
is known (e.g.
responder or non-responder) or be a standard derived from a plurality of
patients whose response is
known.
28
7944618
Date Recue/Date Received 2022-10-28
[000177] In an embodiment, the response threshold is derived from an
unresponsive patient
cancer cell sample and/or a responsive patient cancer cell sample and/or is a
standard derived from a
plurality of unresponsive patient cancer cell samples and/or a plurality of
responsive patient cancer cell
samples.
[000178] In embodiments, a patient RNA integrity value indicative of RNA
integrity and/or a RNA
concentration that is below a response threshold is indicative the cancer is
responding to the radiation
treatment (and/or radiation plus chemotherapeutic agent and/or regimen) and/or
predicts a positive
treatment outcome; and/or a patient RNA integrity value indicative or RNA
integrity and/or a RNA
concentration that is greater than a response threshold is indicative the
cancer is resistant to the radiation
treatment (and/or radiation plus chemotherapeutic agent and/or regimen) and/or
predicts a negative
treatment outcome.
[000179] In an embodiment, the response threshold is a reference or cut-off
RNA integrity value
from subjects with the same or similar tumour type and/or radiation comprising
treatment regimen -
subjects with a RNA integrity value indicative of a RNA integrity and/or a RNA
concentration that is
below or less than the response threshold are predicted (e.g. have an
increased probability) to have a
positive treatment response and/or subjects with a RNA integrity value
indicative of a RNA integrity
and/or a RNA concentration that is above or higher than the response threshold
are predicted to have a
negative treatment response.
[000180] In another embodiment, the response threshold corresponds to the mean
(e.g. average)
RNA integrity value and/or mean RNA concentration for responders and/or non-
responders, for example.
In an embodiment, the mean RNA integrity value is an average of the mean RNA
integrity in cancer cell
samples or ovarian cancer cell samples from cancer subjects respectively that
respond to a radiation
treatment (alone or in combination with a chemotherapeutic agent) and/or the
mean RNA integrity value
is an average of the mean RNA integrity in cancer cell samples from cancer
subjects that do not respond
to treatment. In another embodiment, the response threshold corresponds to a
threshold of high negative
or positive predictive value or high area under the curve by receiver operator
curve analysis or other
probability analysis methods.
[000181] In an embodiment, more than one response threshold can be used. For
example, RNA
integrity values measured using RDA together with or separate from RNA
concentration can be stratified
into multiple zones such as three zones, for example Zone 1: non-responders,
high negative predictive
value, Zone 2: intermediate to include partial responders (some drug effect
but insufficient to achieve
response; this zone may include for example up to 15% of responders), and Zone
3: which is selected
to include for example 85% of responders such as pathological complete
responders and subjects with
increased disease free survival (DES), high positive predictive value. It has
been found for example that
patients treated with chemotherapy and which demonstrate signal induced RNA
degradation falling within
zone 3 have increased DES even if they do not exhibit pCR. The increased DES
is similar to the pCR
population. It is expected that radiation comprising regiments where the RNA
integrity value similarly
29
7944618
Date Recue/Date Received 2022-10-28
identifies patients as falling with zone 3, will have similar DFS benefit.
Other scales and/or formats for risk
assessment based on RNA integrity data could be used, as would be understood
by a person skilled in
the art.
[000182] RDA which can be used for assessing radiation induced RNA integrity
changes, in at
least one embodiment, comprises obtaining at least one electropherogram
dataset corresponding to a
cancer cell sample comprising cellular RNA optionally at a time point before,
during or after the
treatment; determining values for features from the at least one
electropherogram dataset by using two
identifying ranges to accommodate possible shifting of 18S and 28S peaks,
detecting the 18S and 28S
peaks, and calculating the features derived at least in part based on the
located 18S and 28S peaks; and
optionally determining an RDA score based on a combination of the values of
the features.
[000183] In an embodiment, the method comprises obtaining at least one
electropherogram
dataset corresponding to a cancer cell sample comprising cellular RNA at a
time point before, during or
after the treatment; determining values for features from at least two shifted
regions of the at least one
electropherogram dataset, the shifting being due to the treatment; and
optionally determining an RDA
score based on a combination of the values of the features.
[000184] In an embodiment, the RDA, comprises accessing at least one
electropherogram dataset
corresponding to a unique biological sample comprising cellular RNA at a time
point before, during or
after the treatment; determining values for features from the at least one
electropherogram dataset by
using two identifying regions to accommodate possible shifting of 188 and 28S
peaks, detecting the 188
and 28S peaks, and calculating the features derived in part based on the
located 186 and 28S peaks;
and optionally determining an RDA score based on a combination of the values
of the features.
[000185] In another embodiment, the RDA method comprises obtaining at least
one
electropherogram dataset corresponding to a cancer cell sample comprising the
cellular RNA at a time
point; defining an 18S peak and a 28S peak from the at least one
electropherogram dataset; determining
at least one parameter value for both the 188 peak and the 28S peak;
redefining at least one of the 18S
peak and the 28S peak when required according to one or more rules applied to
the at least one
parameter value; determining an 18S peak area and a 28S peak area; and
determining an RDA score
based on at least one of the 18S peak area and the 28S peak area.
[000186] Figure 12 shows different features of the electropherogram that can
be used to calculate
RDI values.
[000187] For example, the features can include one or more of the area of an
188 peak, the area
of a 286 peak, the area of an intermediate banding region, the area of a low
banding region, the area of
one or more band sub-regions, and the total area. The total area is the sum of
the area of the 18S peak,
the 288 peak, the intermediate banding region and the low banding region.
Other features can include
one or more of the width of the 18S peak and the width of the 286 peak. The
area of these peaks and
regions can be calculated in general using well-known mathematical techniques
such as the trapezoidal
rule for numerical integration (Atkinson, 1989).
7944618
Date Recue/Date Received 2022-10-28
[000188] The low banding region can also be divided into the "low C", "low B"
and "low A" sub-
regions or banding regions as demonstrated in Figure 12 for use in some RDA
methods. It has been
found that the low A banding region may contain RNA due to autolytic
degradation as well as due to
other effects and that the low C banding region is an important region in
assessing the effect on RNA due
to various external stimuli such as cytotoxic treatments, for example. It has
also been found that in
response to certain drugs, the RNA starts to spread to the low C banding
region, then to the low B
banding region and then to the low A banding region.
[000189] In an embodiment, the combination of features comprises a ratio of
the 28S peak area to
the 18S peak area, which can be represented by the expressions
"28Sarea/18Sarea" or "28S:18S".
[000190] In an embodiment, the combination of features for RDA comprises a
ratio of the
intermediate banding region area to the addition of the 18S peak area in the
shifted 18S region and the
28S peak area in the shifted 28S region, which can be represented by the
expression "intermediate
area/(18Sarea+28S area)". In an embodiment, the combination of features is
(Intermediate Area+LowC
Area)/(28S Area+18S Area).
[000191] In another embodiment, the combination of features for RDA comprises
a ratio of the low
banding region area to the addition of the 18S peak area in the shifted 18S
region and the 28S peak area
in the shifted 28S region, which can be represented by the expression "low
banding area/(18S+28S)".
[000192] In an embodiment, RDIs are stratified into 3 zones.
[000193] RNA concentration when plotted against RNA integrity values such as
RDI values can
provide additional information. For example, the lower the RNA concentration
in the sample, for a given
RDI the less absolute amount of residual normal RNA present.
[000194] In another embodiment, the response threshold is selected from a mean
maximum RNA
integrity, median maximum RNA integrity, mean RNA integrity, median RNA
integrity, mean minimum
RNA integrity, and median minimum RNA integrity of responders and/or non-
responders.
[000195] In another embodiment, the response threshold is selected from a mean
maximum RNA
concentration, median maximum RNA concentration, mean RNA integrity, median
RNA concentration,
mean minimum RNA concentration, and median minimum RNA concentration of
responders and/or non-
responders.
[000196] In an embodiment, the RNA integrity value is compared to a response
threshold when for
example the response threshold is derived from a control such as a
pretreatment control or untreated
control for example, by one or more probability analysis methods. For example,
the control can be a
subject control, such as a pretreatment sample from the subject. In an
embodiment, where the control is
a pretreatment subject control, a decrease in the RNA integrity value and/or a
RNA concentration
compared to the pretreatment subject control is indicative or cancer
responsiveness and/or positive
treatment outcome. In another embodiment, where the control is a pretreatment
subject control, a
comparable RNA integrity value and/or a RNA concentration - and/or stable RNA
integrity value and/or
31
7944618
Date Recue/Date Received 2022-10-28
RNA concentration- compared to the pretreatment subject control is indicative
of cancer resistance to the
radiation treatment and/or negative treatment outcome post treatment. The
control can be a population
pretreatment control, for example an average, minimum, or maximum RNA
integrity value or RNA
concentration or reference range for two or more subjects with cancer,
optionally breast or ovarian
cancer, prior to treatment.
[000197] In an embodiment, the response threshold predictive of responsiveness
comprises RNA
integrity and/or concentration that is decreased by at least 10%, at least
20%, at least 30%, at least 40%,
at least 50%, at least 60%, at least 70%, at least 80% or at least 90% below a
untreated control such as
a pretreatment value.
[000198] In an embodiment, the RNA integrity value and/or RNA concentration
indicative of
responsiveness and/or positive treatment outcome is decreased at least by 20%,
at least 25%, at least
30%, at least 35%, at least 40%, at least 50%, at least 60%, at least 70%, at
least 80% or at least 90%
below a control such as a pretreatment sample.
[000199] In an embodiment, the RNA integrity value and/or RNA concentration
indicative of
resistance (e.g. or non-responders) and/or negative treatment outcome is
decreased less than 40%, less
than 35%, less than 30%, less than 25%, less than 20%, or less than 10% or is
increased or comparable
compared to a control.
[000200] In another embodiment, the cancer cell sample is obtained after the
patient has received
1, 2, 3, 4, 5, 6, or more doses of radiation treatment. In an embodiment, the
cancer cell sample is
obtained after the patient has completed the radiation treatment and
optionally any adjunct treatment.
[000201] In an embodiment, the patient treatment comprises a dose of a
chemotherapeutic agent.
[000202] In clinical practice radiation can follow chemotherapy. It is likely
that the combination ¨
whether sequential or concurrent would result in increased signal induced RNA
degradation.
[000203] In a further embodiment, the chemotherapeutic agent is selected from
anthracyclines,
taxanes and combinations thereof, preferably wherein the chemotherapeutic
agent comprises epirubicin,
docetaxel sunitinib or combinations thereof. In another embodiment, the taxane
is paclitaxel, docetaxel,
larotaxel, AbraxaneTM, docoxahexaenoic acid-linked paclitaxel, paclitaxel
polyglumex, Ortataxel,
GenexolTM, liposomal-encapsulated paclitaxel, and paclitaxel in a Vitamin E
emulsion.
[000204] In an embodiment, the taxane is docetaxel or paclitaxel.
[000205] In an embodiment, the chemotherapeutic agent is administered in a
chemotherapy
regimen e.g. subsequent to one or more doses of radiation. In another
embodiment, one or more cancer
cell samples comprising cancer cell RNA, are obtained at one or more times
during chemotherapy (after
1, 2, 3, or 4 cycles, and/or any number of cycles or doses) and/or after
completion of the chemotherapy
regimen. In another embodiment, one or more RNA samples correspond to breast
cancer or ovarian
32
7944618
Date Recue/Date Received 2022-10-28
cancer cell samples obtained at one or more times during chemotherapy (after
1, 2, 3, or 4 cycles, and/or
any number of cycles or doses) and/or after completion of the chemotherapy
regimen.
[000206] Cancer cell samples are treated in a manner to minimize RNAse
activity, for example,
cancer cell samples are placed immediately in RNA preservative such as
RNAlater (Qiagen) or other
RNA stabilization reagent or RNA preservative with RNAse inhibitor or flash
frozen for example to -80 C,
for example using liquid nitrogen. A person skilled in the art would be
familiar with the steps taken for
obtaining and storing cancer cell and RNA samples.
[000207] In an embodiment, RNA is isolated from the cancer cell sample and the
RNA integrity
value and/or RNA concentration is measured on the isolated RNA.
[000208] In an embodiment, RNA is isolated/purified from the cancer cell
sample, optionally the
breast cancer cell sample or the ovarian cancer cell sample which is obtained
from the patient. For
example, RNA can be isolated using methods and kits known in the art,
including for example Trizol
based isolations, column based kits such as total RNA extraction columns and
kits. An example of a RNA
isolation method is provided in Example 1. In another embodiment, the method
is performed on pre-
isolated/purified RNA.
[000209] In an embodiment, the method comprises obtaining a cancer cell
sample, optionally a
breast cancer cell sample or an ovarian cancer cell sample after the patient
(or cells) has/have received a
radiation dose, isolating and/or purifying RNA from the cancer cell sample to
provide an isolated/purified
RNA sample comprising cancer cell RNA. The RNA sample is assayed for RNA
integrity and optionally
for RNA concentration.
[000210] The RNA integrity value can be determined for example by any method
that assesses
the state of RNA degradation in cancer cell RNA.
[000211] In an embodiment, the RNA assessed is denatured. In an embodiment,
the RNA
assessed is non-denatured.
[000212] In an embodiment, the RNA integrity value is determined by
calculating a RNA integrity
number (RIN) for example using a method that involves using microfluidics,
microcapillary
electrophoresis, and fluorescent dyes, for example using an Agilent
Bioanalyzer machine, an Experion
Capillary Electrophoresis System with its equivalent RNA Quality Index (RQI),
Nanodrop (Thermo
Scientific, Inc.) or other equivalent systems, such as those manufactured by
Applied Biosystems, Lumex,
or Beckman Coulter Corporation or similar system. In an embodiment, the method
comprises separating
the RNA by microcapillary electrophoresis, detecting RNA for example with
fluorescent dye and
quantitating RNA integrity.
[000213] Recently, microcapillary electrophoresis has been used increasingly
to assess RNA
integrity, particularly since only nanogram quantities of RNA are required.
One such platform, the
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., U.S.A.) uses
microfluidics technology to carry out
electrophoretic separations of RNAs in an automated, reproducible manner
(Mueller, 0. et al.,
33
7944618
Date Recue/Date Received 2022-10-28
Electrophoresis 21(2000) 128-134). The Agilent 2100 Bioanalyzer is now used
in many laboratories for
the assessment of RNA integrity. The development of software for the Agilent
Bioanalyzer allows
calculation of an RNA integrity number (RIN) for each sample after capillary
electrophoresis. (Schroeder,
A. et al., BMC. MoL Biol. 7 (2006) 3; Imbeaud, S. et al. Nucl. Acids Res.
(2005), 33, 6, e56, 1-12). This
software incorporates an algorithm which quantifies the amounts of multiple
RNAs in the
electropherogram of a given RNA sample and assigns a RIN value based on this
assessment.
[000214] For example the Agilent Bioanalyzer uses fluorescent dyes that bind
to nucleic acid to
evaluate RNA concentration and integrity. RNA moves through a separation
channel of a RNA chip, and
intercalating dye binds the RNA. The fluorescence of these molecules is
measured as they pass a
detector.
[000215] In an embodiment, between 20-250 ng of RNA is assessed or any number
in between. In
another embodiment, about 0.5 ng about 1 ng, about 5 ng, about 10 ng, about
2Ong, about 30ng, about
40 ng, about 50 ng is assessed. In another embodiment. The concentration of
the RNA sample is
optionally at least 5 ng/pL, at least 6 ng/pL, at least 7 ng/pL or at least 8
ng/p L.
[000216] In an embodiment, the RNA integrity value is expressed as an RNA
integrity number
(RIN), wherein the RIN comprises a calculation of RNA integrity of multiple
RNAs, preferably wherein the
RIN is calculated using one or more of a RIN algorithm, an analytic
electrophoresis system, or a RNA
chip.
[000217] In an embodiment, a RIN indicative of cancer responsiveness and/or
positive treatment
outcome is less than 4.5, less than 3.5, less than 3, less than 2.5, less than
2, less than 1.5 and/or less
than 1. Accordingly in an embodiment the response threshold is about 4.5,
about 4.0, about 3.9, about
3.8, about 3.7, about 3.6, about 3.5, about 3, about 2.5, about 2, about 1.5
or about 1 and a cancer cell
RIN below the response threshold is indicative of response and/or positive
treatment outcome.
[000218] In an embodiment, a RIN indicative of cancer resistance and/or
negative treatment
outcome is greater than 5, greater than 5.5, greater than 6, greater than 6.1,
greater than 6.2, greater
than 6.3, greater than 6.4, greater than 6.5, greater than 7, greater than 7.5
or greater than 8 and a
cancer cell RIN higher than a response threshold of about 5,about 5.5,about 6,
about 6.1, about 6.2,
about 6.3, about 6.4, about 6.5, about 7, about 7.5 or about 8 is indicative
of resistance and/or negative
treatment outcome.
[000219] In an embodiment, the RNA integrity is measured by calculating a 28S:
18S ribosomal
(rRNA) ratio (e.g. 28S/18S rRNA ratio).
[000220] A 28S rRNA: 18S rRNA ratio can be determined for example by using for
example
denaturing agarose gel systems which can include for example either
formaldehyde and MOPs buffer, or
glyoxal in the loading buffer, to denature the RNA allowing molecules to run
by size. The 28S and 18S
rRNA bands can be visualized for example by ethidium bromide staining or other
more sensitive dyes
such as RiboGreen .
34
7944618
Date Recue/Date Received 2022-10-28
[000221] For example, RNA integrity can be evaluated by visualization of RNA
bands under
ultraviolet light after gel electrophoresis and staining of the gel with
ethidium bromide. Typically, the
intensity values for the 28S and 18S rRNA bands are determined by film
densitometry and a 28S/18S
rRNA ratio computed. RNA is considered of high quality if the 28S/18S rRNA
ratio is about 2.0 or higher.
[000222] In an embodiment, the RNA integrity value and/or the 28S:18s rRNA
ratio is determined
using spectroscopy, for example by assessing UV absorbance at 280:260. In an
embodiment the RNA
integrity value is a ratio of 28S rRNA and 1 8S rRNA.
[000223] In a further embodiment, the RNA integrity value is determined by
assessing the RNA
integrity of a subset of RNAs or fraction of total RNA. In an embodiment, the
RNA is total RNA, ribosomal
RNA or mRNA.
[000224] RNA concentration can be determined by a number of methods including
for example
microcapillary electrophoresis, for example using for example an Agilent
Bioanalyzer machine, an
Experion Capillary Electrophoresis System with its equivalent RNA Quality
Index (RQI), Nanodrop
(Thermo Scientific, Inc.) or other equivalent systems, such as those
manufactured by Applied
Biosystems, Lumex, or Beckman Coulter Corporation or similar system. The RNA
concentration can be
based on UV absorbance, for example by assessing UV absorbance at 280:260.
Either and/or both RNA
concentration can be assessed.
[000225] In an embodiment, the RNA integrity value is measured. In another
embodiment, both
the RNA concentration and the RNA integrity are measured.
[000226] In addition, one or more cancer samples can be assessed. For example
samples can be
obtained before starting radiation treatment, after one or more doses and/or
upon completion.
[000227] In another embodiment, two or more cancer cell samples are obtained.
For example, the
samples can be taken at different time points for example after 1 dose, after
3 doses of radiation and/or
in combination with cytotoxic, optionally chemotherapy treatment.
[000228] In an embodiment, the biopsy is divided into two or more cancer cell
samples and two or
more RNA samples are isolated/purified from the cancer samples and the RNA
integrity value and
optionally the RNA concentration is obtained for each. In an embodiment, an
average RNA integrity value
or RNA concentration of two or more RNA samples is used. In another
embodiment, the highest or
maximum RNA integrity value of the two or more RNA samples is used.
[000229] This approach to monitoring treatment response is expected for
example to permit non-
responding patients (e.g. identified as having moderate to high RNA integrity
after initiation of treatment)
to be switched to other treatments (surgery, other radiation doses, addition
or switch to other drugs)
without completing the remaining cycles of the ineffective regimen. This may
spare patients the toxic
side effects of regimens to which their cancers are not responding.
7944618
Date Recue/Date Received 2022-10-28
[000230] The methods described herein can for example be used to assess and/or
stratify
subjects in a clinical trial.
[000231] A further aspect comprises a method comprising sending a cancer cell
sample optionally
a breast cancer or an ovarian cancer cell sample or RNA sample derived
therefrom to a testing site,
wherein the sample is for example packaged in a RNAse free vessel and
optionally resuspended in a
lysis buffer, RNA isolation and/or stabilization composition optionally
comprising RNAse inhibitors, the
vessel labeled with an identifier permitting, for example, anonymous testing;
and receiving from the
testing site an assessment of the sample's RNA integrity, including a score
(e.g. zone 1, zone 2 or zone
3) or other indicator indicating the risk of treatment failure. The risk
assessment can be provided for
example to a medical practitioner, who will use the risk assessment based on
RNA quality data (in
addition to other data) to decide on the best treatment option to recommend to
his or her patient.
Ill. Kits
[000232] A further aspect includes a kit for use in a method described herein
comprising a RNA
isolating composition and an RNAse free vessel for receiving the tumour and/or
RNA sample, wherein
the vessel is optionally labeled with an identifier optionally permitting for
anonymous testing.
[000233] The above disclosure generally describes the present application. A
more complete
understanding can be obtained by reference to the following specific examples.
These examples are
described solely for the purpose of illustration and are not intended to limit
the scope of the application.
Changes in form and substitution of equivalents are contemplated as
circumstances might suggest or
render expedient. Although specific terms have been employed herein, such
terms are intended in a
descriptive sense and not for purposes of limitation.
The following non-limiting examples are illustrative of the present
disclosure:
Examples
Example 1
Incidence, Mortality, and Treatment of Breast Cancer
[000234] Breast cancer is the most common cancer diagnosis in women, with
22,300 and 5,300
Canadian women being diagnosed with and dying of the disease in 2007,
respectivelyl. Although
treatments have improved both survival and progression-free survival for early
and metastatic breast
cancer patients, those with locally advanced breast cancer (LABC) have
significantly poorer treatment
outcome. LABC is traditionally defined as stage IIB (T3NO) and Stage IIIA/B
from the TMN classification.
Clinically these tumours are greater than 5 cm in size and/or extend beyond
the breast tissue into the
surrounding skin or muscle, with/without matted axillary lymph nodes (N2),
internal mammary nodes (N3)
or ipsilateral supraclavicular lymph node involvement. LABC represents
approximately 10-15% of all
breast cancer cases, and the survival is estimated at 30-42% at 5 years1 a
significant portion of whom
will be living with metastatic disease. However, a small subset of women who
receive neoadjuvant
chemotherapy and achieve a pathologic complete response (pCR), (defined as no
microscopic residual
36
7944618
Date Recue/Date Received 2022-10-28
invasive breast cancer following neo-adjuvant treatment) have a vastly
improved 5 year disease free
survival rate of 87%2 and 5 year overall survival rates of 89% 2 and 90% 3. As
such, pCR rates have
become a surrogate measure for favourable long-term outcomes in trials
involving neoadjuvant
treatment, particularly since this is the only subgroup for which this value
can be measured. The
correlation between improved survival from locally advanced breast cancer and
pCR has been identified
in other studies, mainly using anthracyclines6-8.
Improved Survival of Breast Cancer with the Use of Taxanes
[000235] In order to improve survival from breast cancer, novel cytotoxic
agents have been tested
following or concurrently with anthracycline chemotherapy, notably the
taxanes. Docetaxel is a
microtubule-stabilizing agent which induces cell-cycle arrest at mitosis and
apoptosis9,1 . It has
demonstrated response rates up to 50% in anthracycline-resistant metastatic
breast cancer11-13, and
superior survival when used first-line in randomized studies in the metastatic
setting 14,15. Docetaxel is
most commonly given intravenously every 3 weeks. However, a randomized phase
III study in the
metastatic setting compared docetaxel 100 mg/m2 every 3 weeks, to 35 mg/m2
weekly for 3 of every 4
weeks 16. Although response rates were lower on the weekly arm, there was no
difference in progression-
free survival (5.7 months vs 5.5 months; p = .46) or overall survival (OS)
(18.3 months versus 18.6
months; p = .34). There was higher overall serious toxicity in the Q 3-week
arm, (88.1% versus 55.9%;
p= .0001).
Use of Taxanes in the Neoadiuvant Setting
[000236] Based on its activity in the metastatic setting, docetaxel has been
tested in randomized
trials in early stage breast cancer, and demonstrated superior survival when
added to anthracycline-
based regimens compared to these regimens alone17-18. FEC-D, (fluorouracil 500
mg/m2 IV, epirubicin
100 mg/m2, cyclophosphamide 500 mg/m2 IV every 3 weeks x 3 cycles, followed by
docetaxel 100 mg/m2
IV every 3 weeks x3) is currently one of the most commonly employed regimens
in the adjuvant post-
operative setting.
[000237] Several nonrandomized studies of docetaxel have also shown activity
in the locally
advanced setting either as a single agent, concurrent, or sequentially with
other agent519-23. In order to
determine whether the addition of docetaxel improves outcomes in the pre-
operative setting, several
randomized studies have been conducted. The largest, the NSABP-27, randomized
2411 women with
Tic ¨ T3 NO-N1 disease to receive 4 cycles of AC pre-operatively, versus 4
cycles of AC followed by 4
cycles of docetaxel preoperatively, or 4 cycles of AC preoperatively followed
by surgery and 4 cycles of
post-operative docetaxel. Compared to preoperative AC alone, the addition of
docetaxel significantly
improved clinical complete response (cCR) (40.1% v 63.6%; p < .001), pCR
(13.7% v 26.1%; p < .001)
and proportion of patients with negative nodes (50.8% v 58.2%; p< .001) 24.
With 8.5 years of follow-up,
across all 3 groups, there was no difference in disease free survival (DFS) or
overall survival (OS) (2).
However, in the patients achieving a pCR, there was a significant improvement
in DFS (HR = 0.49, p <
.0001) and OS (HR = 0.36, p < .0001).
37
7944618
Date Recue/Date Received 2022-10-28
[000238] The Aberdeen Breast Group also tested the efficacy of the addition of
docetaxel to an
anthracycline-based regimen in the preoperative setting 25. One hundred and
forty-five women with newly
diagnosed T3, T4 or TxN2 disease received 4 cycles of CVAP (cyclophosphamide
1,000 mg/m2,
doxorubicin 50 mg/m2, vincristine 1.5 mg/m2 and prednisone 40 mg). Those who
achieved a partial or
complete clinical response were then randomized to either 4 more cycles of
CVAP or 4 cycles of
docetaxel (100 mg/m2). Those who did not respond to the initial 4 cycles of
chemotherapy were treated
with docetaxel in a nonrandomized fashion. Intention-to-treat analysis
demonstrated a higher cCR (94% v
66%, p = .03) and pCR (31% v 16% p = .04) with the addition of docetaxel
compared to 4 more cycles of
CVAP. At 38 months median follow-up docetaxel significantly improved DES (90%
v 77%; p = .03) and
OS 97% v 84%; p = .05) 25. A third study, the GEPARDUO study compared AC
(doxorubicin and
cyclophosphamide) for 4 cycles followed by docetaxel for 4 cycles (AC-DOC) to
dose ¨ dense
doxorubicin 50 mg/m2 plus docetaxel 75 mg/m2 every 14 days for 4 cycles with
filgastrim support (ADOC)
preoperatively in 913 women with T1-3 NO-2 breast cancer 27. All endpoints
including clinical response,
pCR and breast-conserving surgery rates were significantly improved with the
sequential AC ¨ T
(doxorubicin and cyclophosphamide followed by a taxane) over the dose dense
arm. Survival endpoints
have not yet been reported.
Use of Radiation To Further Improve Neoadiuvant Chemotherapy Response in
Patients with Locally
Advanced Breast Cancer?
[000239] In spite of the improved outcomes associated with the addition of
taxanes to neoadjuvant
chemotherapy regimens, these gains have been modest. pCR rates for non-
trastuzumb-based regimens
employing both anthracyclines and taxanes are still quite low (15-30%). Since
the achievement of a pCR
in patients is associated with strongly enhanced survival, efforts must be
directed at improving pCR rates
in breast cancer patients, in particular those with locally advanced disease.
One possibility for
improvement of pCR rates for patients with breast cancer is to combine
chemotherapy with radiation
therapy in the neoadjuvant setting. In other tumor sites, a combined modality
approach of local radiation
given concurrently with radio sensitizing chemotherapy or other agent has been
employed in order to
improve outcomes. These include agents such as fluoropyrimidines, platinum and
more recently
taxanes. Postoperative concurrent chemoradiation has long been the standard of
care in locally
advanced rectal cancer. Recent trials comparing preoperative to postoperative
chemoradiation have
demonstrated improved local control with the preoperative approach28,29.
Furthermore, a significant
improvement in pCR and DFS with preoperative chemo/radiotherapy has been
reported 30.
Chemo/radiotherapy is also the standard of care for patients with unresectable
head and neck cancer.
The concurrent approach has demonstrated improvements in organ preservation
and survival over
radiation alone in multiple randomized trials 31-34. In locally advanced non-
small cell lung cancer, several
randomized trials have demonstrated improved local control and survival with
the use of concurrent
versus sequential chemoradiation, most commonly with platinum-based
chemotherapy 35-35. Moreover,
multiple small studies have been done in this patient population adding
docetaxel as a radiosensitizer 33-
44.
38
7944618
Date Recue/Date Received 2022-10-28
[000240] Limited published data exists for the use of neoadjuvant
chemo/radiotherapy in locally
advanced breast cancer. A retrospective review of 44 patients receiving
concurrent chemoradiation with
taxanes for stage I ¨ IV breast cancer has been reported 45. While this study
demonstrated the safety and
feasibility or concurrent chemoradiation with taxanes in locally advanced
breast cancer, response rates
and survival outcomes were not reported.
[000241] A second study of 44 women with stage IIB to III locally advanced
breast cancer has
been reported where patients received twice weekly intravenous paclitaxel 30
mg/m2 for 8 ¨ 10 weeks
concurrent with radiation to total dose of 45 Gy, followed by surgery 46. No
grade 4 toxicities were
observed in the preoperative chemoradiation phase. In the postoperative phase,
the only grade 4 toxicity
was leucopenia (10%). Sixteen percent of patients achieved a pCR, with 18% a
pPR. There was no
association between total dose of preoperative chemotherapy and pCR. A second
study (in abstract
form only) reported on 23 patients receiving 50.4 Gy over 6 weeks, with
paclitaxel 175 mg/m2 day 1, and
5FU 1000 mg/m2/day continuous infusion day 1-3 for 3 cycles every 3 weeks 45.
This was followed by 3
cycles of FEC every 3 weeks, then surgery. Grade 3 toxicities included
radiation dermatitis, esophagitis,
vomiting, mucositis and neutropenia. The clinical CR was 82.6%, and pCR was
52.2%. Overall 2-year
survival is 80.7%.
[000242] Given the above promising findings, a phase II trial evaluating the
FEC-D regimen" was
recently conducted for locally advanced, non-inflammatory breast cancer
patients given in a neoadjuvant
setting, while adding concurrent radiation to the first 6 of 9 weeks of
docetaxel, followed 5 weeks later by
a modified radical mastectomy (which remains the surgical standard of care for
LABC patients). In order
to minimize the side effects of this chemotherapy and optimize its
tolerability with radiation50,51, docetaxel
weekly x 9 weeks was administrated (dose adjusted to 35 mg/m2) rather than q3
weekly x 3 cycles. For
patients whose tumours were Her2-neu gene amplification positive, Herceptin
was given IV q3 weekly for
one year. Toxicities were then evaluated and response rates for this treatment
regimen were assessed.
[000243] Moreover, in a recent clinical trial for patients with locally
advanced breast cancer (NCIC-
CTG-MA.22), the relationship between tumour RNA quality and response to
epirubicin/docetaxel
chemotherapy was assessed. Three image guided core biopsies were obtained from
patients prior to,
during, and immediately after administration of chemotherapy and immediately
flash frozen on dry ice for
subsequent storage in liquid nitrogen. RNA was isolated from the biopsies and
the quantity and quality of
the RNA assessed using an Agilent 2100 Bioanalyzer. RNA quality was quantified
by the bioanlayzer in
terms of an RNA integrity number (RIN), where a RIN of 0 represents completely
hydrolyzed RNA and a
RIN of 10 represents completely undegraded RNA. This study, demonstrated that
chemotherapy
treatment resulted in a dose-dependent reduction in tumour RIN values mid-
treatment and post-
treatment. Moreover, unlike tumour extent/cellularity, low mid-treatment
tumour RIN values were found
to be correlated with the achievement of a pCR in patients post-treatment. All
patients that had a pCR
post-chemotherapy exhibited a reduction in maximum tumour RIN mid-treatment
(mid-treatment in the
above case, was after 3 or 4 cycles of chemotherapy). In the current study of
FEC-D chemotherapy with
concurrent radiation, it was also assessed whether changes in tumour RNA
quantity and quality took
place in response to the regimen.
39
7944618
Date Recue/Date Received 2022-10-28
Methods
Administration of the FEC-D regimen with concurrent radiation
A schema for the above-described study is described in Figure 1. Thirty two
patients with stage III
non-metastatic, non-inflammatory locally advanced breast cancer were treated
with neoadjuvant 5-
Fluoro-uracil, Epirubicin, and Cyclophosphamide (FEC also referred to as CEF)
q3 weekly for 4 cycles
followed by weekly Docetaxel (35 mg/m2) concurrently with regional radiation
(45 Gy with 16 Gy boost in
25 & 5 fractions) for 6 weeks followed by an additional 3 weeks of docetaxel
chemotherapy without
radiation. This was followed by a modified radical mastectomy. Patient and
tumour characteristics were
recorded at baseline and following treatment and clinical response and
treatment-related toxicities noted
(Table1). Image guided serial 14 gauge tumour core biopsies were taken from
the patients pre-, mid-
and post-treatment, and 1 mm3 sections were immediately taken from the
biopsies, immersed in
RNAlaterTM, and stored frozen. In this trial "pre" is before any FEC, mid is
after FEC, MID treatment is
after FEC but before docetaxel with concurrent radiation therapy, and post is
after radiation/docetaxel.
Isolation of RNA from Tumour Core Biopsies
[000244] RNA was isolated from image-guided tumour core biopsies of patients
pre-, mid-, and
post-treatment using Qiagen miRNeasy Mini kits, following a modification of
the protocol published on
the manufacturer's we bsite, http://www1 .giag en.co Write rature/h and boo
ks/ literature.aspx?id=1000291 .
Biopsies were cut into several sections for various assays, with the section
used for RNA integrity
analysis placed in RNAlater.The biopsies in RNAlater were immediately dropped
in 0.5 ml of RLT buffer
containing 3-ME (10 pl into 1 ml) in a 1.5 ml tube. The biopsies in RLT buffer
were homogenized with a
Coreless motor homogenizer for 5 min (from the Kontes Glass Company). The
lysate was then passaged
at least 5 times through a 20-gauge needle (0.9 mm diameter) fitted to an
RNase-free syringe. The
sample was then centrifuged at high speed in a refrigerated microfuge at 4 C
for 3 minutes, with transfer
of the supernatant to a new tube. One volume (500 pl) of 70% ethanol was then
added to the
supernatant and the sample mixed well by repeated pipetting. If some lysate
was lost during
homogenization, then the volume of ethanol was adjusted accordingly. Visible
precipitates formed after
the addition of ethanol in some samples did not affect the RNA isolation
procedure. A maximum of 700 p1
of the sample, including any precipitate, were added to a Qiagen mini column
and placed in a 2 ml
collection tube. The column was centrifuged for 15 s at 8000 x g W10,000 rpm)
and the flow-through
discarded. The remainder of the sample was then added to the column and the
column centrifuged
again. From this point forward, the column was then washed twice in RPE buffer
and dried by
centrifugation as per the manufacturer's protocol. The RNA was then eluted
from the column in 30 pl of
RNase-free water and the eluate reapplied and eluted from the column to
increase the yield and
concentration of the RNA obtained.
7944618
Date Recue/Date Received 2022-10-28
Assessment of RNA quality using an Aqilent 2100 Bioanalyzer
[000245] The above RNA samples were applied to RNA 6000 Nano LapchipsTM
(purchased from
Agilent Biotechnologies, Inc.) and subjected to capillary electrophoresis
using an Agilent 2100
Bioanalyzer. The protocol followed was identical to that described in the
company's technical brochure
for the Agilent 2100 Bioanalyzer, available
at:
http://www.chem.agilent.com/scripts/LiteraturePDF.asp?iWHID=36225. The amount
and quality (RI N
value) of RNA from each core biopsy was then determined by the Bioanalyzer.
Results
[000246] Clinical Responses and Toxicities to FEC-D chemotherapy with
concurrent radiation
While 30 of the 32 patients (94%) completed the treatment protocol described
above, patients
experienced significant toxicities. Twenty seven patients (84%) had grade 3 or
greater toxicities, including
grade 3 resolving pneumonitis (6 patients), grade 3 dermatitis (6 patients)
and one treatment-related
death. As shown in Table 1, eight of these patients (25%) exhibited a
pathologic complete response
(pCR) to treatment, which is approximately twice the Ontario pCR rate for
locally advanced breast
cancer. Moreover, at a mean 21 months of follow-up, the relapse-free survival
rate was 100% in the pCR
cohort and 65% among partial responders (PRs). This suggests that the regimen,
while exhibiting strong
toxicity, appears to enhance the pCR and survival rate for locally advanced
breast cancer. Tumours that
exhibited pCRs were distributed almost equally amongst the basal (2 of 5
tumours = 40%) , Her2 (3 of 3
tumours = 100%), and luminal B (3 of 6 tumours = 50%) subtypes. No pCRs were
found among the 11
patients with luminal A tumours (0%). While the numbers are small, the data
suggests that FEC-D
regimen with concurrent radiation appeared able to induce pCRs across a
variety of breast cancer
subtypes.
41
7944618
Date Recue/Date Received 2022-10-28
ID AGE Base Stage Path Response Path Stage ER PR Her2 Subtype-
Like Mean 2.5 Yr F/U
1 62 Stage IHA PR Stage 1 Negative Negative
Positive .Her2 .Died
2 38 Stage IIB CR Stage 0 Negative Negative
Positive Her2 Alive No Disease
._ .
3, 26 Stage IIIA CR Stage 0 Negative Negative
Negative Basal Alive No Disease
_.......
41 58'Stage IIIA CR Stage 0 Positive Positive
Positive Luminal B Alive No Disease
_.,_. ,
1 5 43 Stage IIIA Stable Stage IIIA Positive
Positive Positive Luminal B Alive No Disease
, 6 52 Stage WA Stable Stage IIB Positive
Positive Positive Luminal B Alive No Disease
7 49 Stage IIIA CR Stage 0 Positive Positive
Positive Luminal B Alive No Disease
_..
1 8 63 Stage MA Prog Stage IV Negative Negative
Negative Basal Died
,
I* 91 48 Stage IIIA PR Stage 1113
Positive Positive Negative Luminal A Alive No Disease
......_ ........
¨10 61 Stage IIIA PR __ StagellA Positive Positive
Negative Luminal A Died
11 39 Stage IIB PR Stage IIA Positive Positive
Negative Luminal A Died
12 47 Stage IIB . PR Stage I Positive
Positive õNegative Luminal A Alive No Disease _....
131 43 Stage IIB , CR Stage 0 ___ Negative
Negative Positive Her2 Alive No Disease
,
141 49 Stage IIIA PR Stage IIA Negative Negative
Negative Basal Died
15 64 Stage IIIA PR Stage IIIA .Positive Negative
Negative Luminal A Died .
16 34 Stage IIIC PR Stage I Negative Negative
Negative õBasal Alive No Disease
17 40 Stage IIB PR Stage I Positive Positive
Negative Luminal A Alive No Disease
. 18, 58 Stage IIIC .
PR Stage IIA Positive
,Positive Positive Luminal B Alive No Disease
. 191 42 Stage IIIA Stable Stage IIA Positive
Positive Negative Lumina' A Alive No Disease
. 20 53 Stage IIIB CR Stage 0 Negative Negative
Negative Basal Alive No Disease
. 21 44 Stage IIB PR Stage 1 Positive Positive
Positive Luninal B Alive No Disease
22. 45 Stage IIB PR Stage IIA Positive Negative
Negative Luminal A Alive No Disease
231 57 Stage IIIA CR Stage 0 Positive Negative
Positive Luminal B Alive No Disease
'
. 24. 60 Stage IIB PR Stage IIB Positive
Positive Negative Lumina] A Alive WITH Disease
251 50 Stage IIIA PR Stage IIB Negative Negative
Negative Basal Alive WITH Disease
26 44 Stage IIIB PR 1Stage IIA Positive Positive
Negative Luminal A Alive No Disease
27 45 Stage IRA PR 'Stage ILA Positive Positive
Negative Luminal A Alive No Disease
281 62 Stage IIIA PR Stage IIIA Positive Negative
Positive Lumina' B Alive No Disease
291 51 Stage IIIB PR Stage IIA Positive Positive
,Positive Luminal B Alive No Disease
30 58 Stage IRA NA Died in Tx Positive Positive
Positive Luminal B .Died
31 31 Stage IIIC PR Stage IIA Positive Positive
Positive Luminal B Alive No Disease
32, 62 Stage IRA PR Stage IIIA ............... Positive Positive
Negative Luminal A Alive No Disease
[000247] Table 1 Data on patient characteristics prior to and post-treatment
in the London Clinical
Study, including age, baseline nodal status and stage, completion of
treatment, level of pathologic
response, post-treatment nodal status and stage, pre-treatment receptor
expression status, predicted
tumour subtype and degree of toxicity experience by patients. Y= yes, N= no,
PR=partial response,
CR=complete response, Stab=stable disease, Prog=progressive disease.
42
7944618
Date Recue/Date Received 2022-10-28
r-
TOXICITY
Category Quantity
.... ....
Radiation Pneumonitis 25%
Radiation Dermatitis ........................... 25%
ARDS Death N=1
2.5 YEAR SURVIVAL
Patients With pCR N=7, 100%
Patients Without pCR N=24, 62%
pCR BY TUMOUR SUBTYPE
ER+/PR+or-/Her2- (Lumi nal A) N = 0/12 =0%
ER-/PR+or-/Her2+ (Lumina! B) N = 3/10 = 30%
ER-/PR-/Her2+ (Her2) N = 2/3 = 66%
ER-/PR-/Her2- (Basal) N = 2/6 = 33%
[000248]
Table 2: Effects of FEC chemotherapy followed by docetaxel treatment with
concurrent radiation therapy in LABC patients. Toxicities included radiation
pneumonitis and dermatitis
and one death during treatment from acute respiratory distress. Syndrome
(ARDS). Two year survival
post-treatment was higher for patients achieving pCR. pCRs occurred in
patients with tumours of the
Her2, luminal B, and basal subtypes, but were absent in luminal A tumours.
43
7944618
Date Recue/Date Received 2022-10-28
Changes in Tumour RNA Content In Response to FEC chemotherapy followed by
docetaxel
chemotherapy with concurrent radiation treatment
[000249] It was then assessed whether, similar to the NCIC-CTG-MA.22 clinical
trial, changes in
tumour RNA quality or quantity could be observed during or in response to
treatment and whether low
RNA quality was associated with a strong clinical response upon the completion
of treatment (pCR).
Figure 2 illustrates the RNA concentration values for all patient biopsies
isolated prior to treatment, in the
middle of treatment, and post-treatment. This plot reveals that there was some
significant variability in the
quantity of RNA isolated from the biopsies throughout treatment, including pre-
treatment biopsies. This
suggests possible variations in the preservation of RNA in the collected
biopsies. In addition, the data
suggests little difference in RNA concentration between pre-treatment biopsies
and biopsies collected
after FEC chemotherapy (mean tumour RNA concentration of 50.0 15.1 and 50.0
11.9 ng/ 1,
respectively). In contrast, the mean tumour RNA concentration fell
significantly after the completion of the
FEC-D regimen with concurrent radiation and was 10.6 2.1
nanograms/microliter. These findings
suggest that the FEC chemotherapy alone is insufficient to induce reductions
in tumour RNA
concentration, but upon treatment with concurrent radiation therapy and
docetaxel, tumour RNA content
falls dramatically. Despite this treatment effect, there were no significant
differences in tumour RNA
concentration were observed amongst patients that exhibited a pathologic
complete response post-
treatment (pCR), patients that exhibited a partial response to treatment (PR),
and patients with stable or
progressive disease (SD or PD) post-treatment (Figure 3). Changes in Tumour
RIN Values In Response
to FEC chemotherapy followed by docetaxel chemotherapy with concurrent
radiation treatment.
[000250] To assess changes in tumour RNA content in during FEC-D chemotherapy
with
concurrent radiation treatment, all samples that were noted as "insufficient
signal" for mathematical
analysis were omitted An insufficient signal is one with no detectable peaks
above background. As
shown in Figure 4A, in the three samples post-FEC chemotherapy but before
docetaxel/radiation
treatment which achieved a pCR at the end of treatment, 2 out of 3 samples
demonstrated RIN values
indicative of high RNA integrity or minimal RNA degradation (e.g. RIN > 7
which is comparable to
baseline levels). One patient sample had a very low RIN value suggestive of
significant loss of RNA
integrity (RIN value = n/a or 0). In the samples from non-responding patients
(patients who did not
achieve pCR post-treatment), little effect of FEC treatment on tumour RNA
quantity or integrity could be
observed when compared to pre-treatment samples.
[000251] When tumour RNA integrity was assessed after both FEC chemotherapy
and
docetaxel/radiation treatment, only two samples from patients that achieved a
pCR post-treatment had
sufficient RNA for mathematical analysis (Figure 4B). For these two samples,
both had RIN values of n/a
or zero, indicative of very strong loss of RNA integrity. These two responders
to treatment were strongly
distinct from nonresponders based on RNA concentration and RIN values. The low
RIN values are
indicative of loss of normal (e.g. non-fragmented) RNA. In the nonresponders,
a wide range of RIN
values are noted which is indicative of a spectrum of change in tumour RNA
from highly fragmented to
highly intact. These results suggest that loss of RNA integrity occurred with
radiation and docetaxel
44
7944618
Date Recue/Date Received 2022-10-28
resulting in decreased RIN values and a loss in RNA concentration. This loss
of RNA integrity correlated
with a strong response to treatment (pCR). High RNA concentration and high RIN
is suggestive of non-
response based on the clinical data.
Example 2
Changes in Tumour Cell RNA integrity In Vitro in Response to Radiation with or
without Docetaxel
Treatment
[000252] There are genomic similarities between ovarian and breast cancers
(Nature 490:67-70, 4
October, 2012) and may show similar responses. To this end, we looked at an
ovarian cancer line for its
RNA response to radiation therapy.
[000253] Experiments were conducted to assess whether radiation with or
without docetaxel
treatment could induce changes in cellular RNA concentration and/or RIN values
in A2780 ovarian
tumour cells. As shown in Figure 5A, radiation treatment induced a dose-
dependent change in A2780
cell RNA content, with 10 Gray of radiation inducing almost a 5-fold reduction
in RNA concentration
levels. Interestingly, pre-incubation of cells with 0.1 p.M docetaxel further
reduced cellular RNA
concentration by an additional 6.5-fold, almost eliminating all RNA from the
cells. In contrast under the
tested conditions, radiation doses up to 5 Gray had no effect on cellular RNA
integrity, as no changes in
RIN values were observed compared to samples prior to radiation treatment
(Figure 5B). If cells were
pre-incubated for 24 hours with 0.1 1.1M docetaxel prior to 10 Gray radiation
treatment, the cellular RIN
value decreased to 75% of pre-treatment values. There were additional
decreases in RIN when radiation
and chemotherapy were used together concurrently. Taken together, these
findings suggest that
radiation can induce reductions in cellular RNA content and reductions in RNA
integrity. Moreover, the
combination of both radiation and docetaxel treatment induces even stronger
reductions in both cellular
RNA content and RNA integrity. These observations are consistent with those
described above for
tumours of patients with locally advanced breast cancer, in particular for the
combined therapy post-
treatment, where RNA concentration fell dramatically in response to FEC-D
chemotherapy with
concurrent radiation therapy and where differences between between responding
patients (pCR) and
non-reponding patients (no pCR) appeared to be observed post-treatment.
[000254] It
was then examined whether similar observations could be observed in vitro for
specific breast tumour cell lines. As shown in Figure 6A, a change in RNA
concentration of about 7%
was observed for SKBR3 breast tumour cells in culture when treated with up to
10 Gray of radiation. If
cells were pre-treated with 0.1 t.iM docetaxel prior to 10 Gray radiation, a 5-
fold reduction in cellular RNA
concentration was observed (19% of untreated cells). RNA concentration
decreases 2 fold (or more) in
BT-20, A2780 and MB-468 cells with radiation only. These findings suggest that
in breast tumour cells in
vitro (and possibly in the tumours of breast cancer patients), some changes in
cellular RNA concentration
may be observed by radiation treatment alone. Concurrent docetaxel treatment
with radiation therapy
dramatically reduces cellular RNA content, consistent with the observed
findings for patients with locally
advanced breast cancer treated with FEC-D chemotherapy followed by radiation
treatment. Similar
7944618
Date Recue/Date Received 2022-10-28
findings were observed when the effects of the above treatments on RIN values
were examined in
SKBR3 (HER2 +, ER -) cells (Figure 6B). Measurable (9%) reductions in cellular
RIN values was
observed in response to radiation if cells were pre-treated with docetaxel.
[000255]
The effects of the above treatments on MCF-7 (ER +, HER2 -) cells (Figures 7A
and 7B) were examined, findings were similar to those of SKBR3 cells, with 4%
and 73% reductions in
RNA levels observed for the 10 Gray and 10 Gray/0.1 i.LIVI docetaxel
treatments, respectively. Similarly,
no significant reduction in cellular RIN values were observed for MCF-7 cells,
even at the highest
radiation dose (10 Gray). Moreover, only a 4% reduction in cellular RIN values
were observed when
cells were pre-treated with 0.1 1.1M docetaxel prior to the 10 Gray radiation.
Example 3
[000256] A2780 cells
were treated with increasing doses of Epirubicin with and without
10Gy radiation.A2780 cells were plated into 6 cm plates in 4 ml of RPM!
culture medium. The plates were
placed in the incubator and the cells were allowed to adhere overnight. The
next day, the medium was
removed from all plates and new media containing the appropriate epirubicin
concentration (1 pM, 5 pM,
nM, 50 nM, and 100 nM) was added to the plates. Cells were pre-treated for 24
h with epirubicin prior
20 to receiving radiation treatment. The following day (24 h post epirubicin
drug-treatment), the cells were
given 10 Gy of x-ray radiation treatment (300 kV, 9 mA) in approximately 4.3
min of x-ray exposure using
a RS320 Irradiation System (Gulmay Medical) The cells were returned to the
cell culture incubator, and
RNA was extracted from the cells 96 h post-radiation treatment (which
corresponds to 120 h post-
epirubicin treatment).
[000257] Figure 8A
shows RIN values versus Epirubicin dose. A loss in RIN value is seen
with radiation at low doses of Epirubicin.
[000258]
Figure 8B shows RDI values vs Epirubicin dose. RDI values increase with
radiation at low drug doses.
[000259]
Figure 8C shows RNA concentration vs. Epirubicin Dose and demonstrates an
increased loss in RNA concentration with radiation at low drug doses.
[000260]
Similar experiments were conducted with MCF-7 breast cancer cells. MCF-7
cells were treated with increasing doses of Epirubicin with and without 10Gy
radiation. MCF-7 cells were
plated into 6 cm plates in 4 ml of DMEM culture medium. The plates were placed
in the incubator and the
cells were allowed to adhere overnight. The next day, the medium was removed
from all plates and new
media containing the appropriate epirubicin concentration (1 pM, 5 pM, 20 nM,
50 nM, and 100 nM) was
added to the plates. Cells were pre-treated for 24 h with epirubicin prior to
receiving radiation treatment.
The following day (24 h post epirubicin drug-treatment), the cells were given
10 Gy of x-ray radiation
treatment (300 kV, 9 mA) in approximately 4.3 min of x-ray exposure using a
RS320 Irradiation System
(Gulmay Medical) The cells were returned to the cell culture incubator, and
RNA was extracted from the
cells 96 h post-radiation treatment (which corresponds to 120 h post-
epirubicin treatment).
[000261]
Figure 9A shows RIN value vs. Epirubicin Dose and demonstrates a loss in RIN
value when combined with radiation.
46
7944618
Date Recue/Date Received 2022-10-28
[000262] Both MCF-7
and SkBr3 cells had a 45% increase in RDI with 2Gy of radiation
only. RDI is more sensitive and the RNA degradation effect is less evident as
a change in the RIN value.
[000263]
Figure 9B shows RDI value vs. Epirubicin Dose and demonstrates an increase in
RDI value with radiation up to 100 uM Epirubicin. Figure 9C shows RNA
concentration vs. Epirubicin
Dose and demonstrates a loss in RNA concentration with increasing doses of
Epirubicin. RDI is more
sensitive than RIN in measuring radiation induced effects.
[000264] In
another experiment, A2780 cells were treated with 5 nM Docetaxel, 10 Gy
Radiation and 10 Gy Radiation + 5 nM Docetaxel for 72 hours or 96 hours. The
results demonstrate that
signal induced RNA degradation is time-dependent.
[000265]
Figure 10A provides the mean RI N value versus drug and radiation treatment
(n=3). Error bars represent standard deviation. Figure 10A shows an increased
loss in RIN value with
radiation + Docetaxel at 72 hr.
[000266]
Figure 10B provides the mean RDI value versus Docetaxel and radiation
treatment (n=3). The results suggest an increase in RDI value with
radiation+Docetaxel compared with
Docetaxel alone at 96 hr. Error bars represent standard deviation.
Example 4
[000267]
Electropherogram data obtained from patients in Example 1 was reanalyzed by
calculating RDI values. The RDI values were calculated using features
(Intermediate Area+LowC
Area)/(28S Area+18S Area).
[000268]
Figure 11 plots RNA concentration versus RDI values for A) mid-treatment
(before radiation) and B) post treatment samples (after radiation).
[000269]
Based on the above, an RDI value of 3 or less may indicate non-response to
radiation. For example, Figure 11 b shows that post-treatment samples which
had a RDI vaule of 3 or less
are non-responders to the radiation/docetaxel combination. High RNA
concentration and low RDI are
suggestive of non-response post treatment.
Example 5
[000270]
A2780 ovarian cancer cells were treated with radiation of 2 to 10 Gray using a
Gulmay R5320 Irradiation System and subsequently harvested after 24, 48 and 72
hr. RNA was
isolated and run on an Agilent Bioanalyzer. Analysis of the electropherogram
demonstrated that an
increase in the ratio Intermediate Area/(28S+18S Areas) at 72 hours is
detectable in radiation treated
cells. For example, cells treated at 10 Gray for 72 hours show radiation
induced RNA degradation in the
intermediate region and the low banding region, but not much difference in the
area of the
electropherogram where the autolysis peak resides. Radiation induced RNA
degradation is evident at
72 hrs at both dose levels.
[000271]
While the present application has been described with reference to what are
presently considered to be the preferred examples, it is to be understood that
the application is not
limited to the disclosed examples. To the contrary, the application is
intended to cover various
modifications and equivalent arrangements included within the spirit and scope
of the appended claims.
47
7944618
Date Recue/Date Received 2022-10-28
[000272] All publications, patents and patent applications are herein
incorporated by
reference in their entirety to the same extent as if each individual
publication, patent or patent
application was specifically and individually indicated to be incorporated by
reference in its entirety.
Specifically, the sequences associated with each accession numbers provided
herein including for
example accession numbers and/or biomarker sequences ( e.g. protein and/or
nucleic acid) provided in
the Tables or elsewhere, are incorporated by reference in its entirely.
48
7944618
Date Recue/Date Received 2022-10-28
CITATIONS FOR REFERENCES REFERRED TO IN THE SPECIFICATION
1. Canadian Cancer Society. Canadian Cancer
Statistics. http://www cancer
ca/vgn/images/portal/cit_86751114/10/34/614137951cw_library_INYNTK_Bladder_Punj
abi2005 pdf [
2008 Available from: URL:www.cancer.ca
2. Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K et al. Clinical
course of breast
cancer patients with complete pathologic primary tumor and axillary lymph node
response to
doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol 1999; 17(2):460-469.
3. Formenti SC, Dunnington G, Uzieli B, Lenz H, Keren-Rosenberg S, Silberman H
et al. Original p53
status predicts for pathological response in locally advanced breast cancer
patients treated
preoperatively with continuous infusion 5-fluorouracil and radiation therapy.
Int J Radiat Oncol Biol
Phys 1997; 39(5)1059-1068.
6. Scholl SM et al. Breast tumours response to primary chemotherapy
predicts local and distant control
as well as survival. Eur J Ca 1995; 31A: 1969-1995.
7. ChoIlet P et al. Clinical and pathological response to primary chemotherapy
in operable breast
cancer. Eur J Ca 1997; 33: 862-866.
8. Gazet JC et al. Assessment of the effect of pretreatment with neoadjuvant
therapy on primary breast
cancer. Br J Ca 1996; 73: 758-762.
9. Bissery MC et al. Experimental antitumor activity of Taxotere (RP 56976,
NCS 628503) a Taxol
analogue. Ca Res 1991; 51: 4845-4852.
10. Ganansia-Leymarie Vet al. Signal transduction pathways of taxane-induced
apoptosis. Curr Med
Chem Antica Ag 2003; 3: 291-306.
11. Valero Vet al. Phase II trial of docetaxel: a new highly effective
antineoplaslic agent in the
management of patients with anthracycline-resistant metastatic breast cancer.
JCO 1995; 13: 2886-
2894.
12. Ravdin PM et al. Phase II trial of docetaxel in advanced anthracycline-
resistant or anthracenedione-
resistant breast cancer. JCO 1995; 13: 2879-2885.
13. Chan Set al. Prospective randomized trial of docetaxel versus doxorubicin
in patients with metastatic
breast cancer. The 303 Study Group. JCO 1999; 17: 2341-2354.
14. Nabholtz JM et al. Docetaxel and doxorubicin compared with docetaxel and
cyclophosphamide as
first-line chemotherapy for metastatic breast cancer: results of a randomized,
multicenter phase III
trial. JCO 2003; 21: 968-975.
15. O'Shaughnessy J et al. Superior survival with capecitabine plus docetaxel
combination therapy in
anthracycline-pretreated patients with advanced breast cancer; phase III trial
results. JCO 2002; 20:
2812-2823.
16. Rivera E et al. Phase 3 study comparing the use docetaxel on an every-3-
week versus weekly
schedule in the treatment of metastatic breast cancer. Cancer 2008; 112: 1455-
1461.
17. Roche H et al. Sequential adjuvant epirubicin-based and docetaxel
chemotherapy for node-positive
breast cancer patients; the FNCLCC PACS 01 trial. JCO 2006; 24: 5664-5671.
18. Martin M et al. Adjuvant docetaxel for node-positive breast cancer. NEJM
2005; 352: 2302-2313.
19. Amat Set al. Induction chemotherapy in operable breast cancer: high
pathological response rate
induced by docetaxel. Proc Am Soc Clin Onc 1999; 18: 79a, abstract 297.
20. Tubiana-Hulin M et al. Phase 11 trial combining docetaxel (D) doxorubicin
(DOX) as neoadjuvant
treatment in patients (Pts) with operable breast carcinoma (BC). Proc Am Soc
Clin Onc 2000; 19:
127a, abstract 492.
21. Limentani SA et al. Phase 11 study of doxorubicin and docetaxel as
neoadjuvant therapy for women
with stage IIB or III breast cancer. Proc Am Soc Clin Onc 2000; 19: 131a,
abstract 511.
22. Teston L et al. Dose-dense chemotherapy with sequential doxorubicin (D)
and docetaxel (Dt) for intial
treatment of operable and inoperable stage 11-111b breast cancer. Proc Am Soc
Clin Onc 2000; 19:
134a, abstract 524.
23. VVynendale Wet al. Neoadjuvant chemotherapy with sequential doxorubicin
(DOX) and docetaxel
(DOC) in locally advanced breast cancer (LABC): a pilot study. Proc Am Soc
Clin Onc 1999; 18:
106a, abstract 389.
24. Bear HD et al. the effect on tumor response of adding sequential
preoperative docetaxel to
preoperative doxorubicin and cyclophosphannide: preliminary results from
National Surgical Adjuvant
Breast and Bowel Project protocol B-27. JCO 2003; 21: 4165-4174.
25. Smith IC et al. Neoadjuvant chemotherapy in breast cancer significantly
enhanced response with
docetaxel. JCO 2002; 20: 1456-1466.
26. Hutcheon AW et al. Improvements in survival in patients receiving primary
chemotherapy with
docetaxel for breast cancer: a randomized controlled trial. Br Ca Res Tr 2001;
69: 298.
49
7944618
Date Recue/Date Received 2022-10-28
27. Von Minckwitz G et al. Doxorubicin with cyclophosphamide followed by
docetaxel every 21 days
compared with doxorubicin and docetaxel every 14 days as preoperative
treatment in operable breast
cancer: the GEPARDUO study of the German Breast Group. JCO 2005; 23: 2676-
2685.
28. Sauer R et al. Preoperative versus postoperative chemoradiotherapy for
rectal cancer. NEJM 2004;
351: 1731-1740.
29. Bosset JF et at. Chemotherapy with preoperative radiotherapy in rectal
cancer. NEJM 2006; 355:
1114-1123.
30. Rode! C et al. Prognostic significance of tumor regression after
preoperative chemoradiotherapy for
rectal cancer. JCO 2005; 23: 8688-8696.
31. Adelstein DJ et al. An Intergroup phase III comparison of standard
radiation therapy and two
schedules of concurrent chemoradiotherapy in patients with unresectable
squamous cell head and
neck cancer. JCO 2003; 21: 92-98.
32. Denis F et al. Final results of the 94-01 French Head and Neck Oncology
and Radiotherapy Group
randomized trial comparing radiotherapy alone with concomitant
radiochemotherapy in advanced-
stage oropharynx carcinoma. JCO 2004; 22: 69-76.
33. Forastiere AA et at. Concurrent chemotherapy and radiotherapy for organ
preservation in advanced
laryngeal cancer. NEJM 2003; 349: 2091-2098.
34. Posner MR et at. Cisplatin and Fluorouracil alone or with docetaxel in
head and neck cancer. NEJM
2007; 357: 1705-1715.
35. Furuse K et at. Phase III study of concurrent versus sequential thoracic
radiotherapy in combination
with mitomycin, vindesine and cisplatin in unresectable stage III non-small
cell lung cancer. JCO
1999; 17: 2692-2699.
36. Curran Wet at. Phase III comparison of sequential versus concurrent chemo-
radiation for patients
with unresected stage III non-small cell lung cancer (NSCLC): report of
Radiation Oncology Group
(RTOG) 9410. Lung Ca 2003; 29 (suppl 1): 93, abstract 303.
37. Pierre F et al. a randomized phase III trial of sequential chemo-
radiotherapy versus concurrent
chemo-radiotherapy in locally advanced non-small cell lung cancer (NSCLC)
(GLOT-GFPC NPC 95-
01 study). Proc Am Soc Clin Onc 2001; 20: 312a, abstract 1246.
38. Zatloukal PV et at. Concurrent versus sequential radiochemotherapy with
vinorelbine plus cisplatin
(V-P) in locally advanced non-small cell lung cancer. A randomized phase ll
study. Proc Am Soc Clin
Onc 2002; 21: 290a, abstract 1159.
39. Mauer AM et al. Phase I study of docetaxel with concomitant thoracic
radiation therapy. JCO 1998;
16: 159-164.
40. Mudad R et al. Concomitant docetaxel, cisplatin and radiation (XRT) in the
treatment of locally
advanced non-small cell lung cancer (NSCLC): a phase I study. Proc Am Soc Clin
Onc 2000; 19:
544a.
41. Koukourakis MI et al. Weeky docetaxel and concomitant boost radiotherapy
for non-small cell lung
cancer. A phase I/II dose escalation trial. Eur J Ca 1998; 34: 838-844.
42. Ramlan R et at. Randomized phase II study evaluating the feasibility of
thoracic radiotherapy with or
without weekly docetaxel (Taxotere@) following induction chemotherapy with
cisplatin and docetaxel
in unresectable stage III A-B non-small cell lung cancer. ESMO Congress 2002;
Poster 491.
43. Wu HG et al. Phase I study of weekly docetaxel and cisplatin concurrent
with thoracic radiotherapy in
stage III non-small cell lung cancer. Int J Rad Onc Bio Phys 2002; 52: 75-80.
44. Onishi H et al. Concurrent two-dimensional radiotherapy and weekly
docetaxel in the treatment of
stage III non-small cell lung cancer: a good local response but no good
survival due to radiation
pneumonitis. Lung Ca 2003; 40: 79-84.
45. Bellon JR et at. Concurrent radiation therapy and paclitaxel or docetaxel
chemotherapy in high-risk
breast cancer. Int J Rad Onc Bio Phys 2000; 48: 393-397.
46. Formenti SC et at. Preoperative twice-weekly paclitaxel with concurrent
radiation therapy followed by
surgery and postoperative doxorubicin-based chemotherapy in locally advanced
breast cancer: a
phase I/II trial. JCO 2003; 21: 864-870.
48. Brewer-Goubely YP et al. Neoadjuvant concurrent chemoradiotherapy (CT-RT)
with paclitaxel
(TAXOL) and 5-fluorouracil (5-FU) followed by epirubicin-cyclophosphamide
(FEC) and surgery in
patients (Pts) with locally advanced breast cancer (LABC). Proc Am Soc Clin
Onc 2001; 20, abstract
1815.
49. Roche H, Fumoleau P, Spielmann M, Canon JL, Delozier T, Serin D et at.
Sequential adjuvant
epirubicin-based and docetaxel chemotherapy for node-positive breast cancer
patients: the FNCLCC
PACS 01 Trial. J Clin Oncol 2006; 24(36):5664-5671.
50. Tabernero J, Climent MA, Lluch A, Albanell J, Vermorken JB, Barnadas A et
at. A multicentre,
randomised phase ll study of weekly or 3-weekly docetaxel in patients with
metastatic breast cancer.
Ann Oncol 2004; 15(9):1358-1365.
7944618
Date Recue/Date Received 2022-10-28
51. Rivera E, Mejia JA, Arun BK, Adinin RB, Walters RS, Brewster A et al.
Phase 3 study comparing the
use of docetaxel on an every-3-week versus weekly schedule in the treatment of
metastatic breast
cancer. Cancer 2008; 112(7):1455-1461.
52. A.Schroeder, 0.Mueller, S.Stocker, R.Salowsky, M.Leiber, M.Gassmann,
S.Lighffoot, W.Menzel,
M.Granzow, and T.Ragg, The RIN: an RNA integrity number for assigning
integrity values to RNA
measurements, BMC.Mol.Biol. 7 (2006) 3.
53. A.M.Parissenti, J.A.Chapman, H.J.Kahn, B.Guo, L.Han, P.O'Brien,
M.P.Clemons, R.Jong, R.Dent,
B.Fitzgerald, K.I.Pritchard, L.E.Shepherd, and M.E.Trudeau, Association of low
tumor RNA integrity
with response to chemotherapy in breast cancer patients, Breast Cancer
Res.Treat. 119 (2010) 347-
356.
54. C.Cera, G.Palu, S.M.Magno, and M.Palumbo, Interaction between second
generation anthracyclines
and DNA in the nucleosomal structure, Nucleic Acids Res. 19 (1991) 2309-2314.
55. S.Spadari, G.Pedrali-Noy, F.Focher, A.Montecucco, T.Bordoni, C.Geroni,
F.C.Giuliani, G.Ventrella,
F.Arcannone, and G.Ciarrocchi, DNA polymerases and DNA topoisomerases as
targets for the
development of anticancer drugs, Anticancer Res. 6 (1986) 935-940.
56. N.R.Bachur, F.Yu, R.Johnson, R.Hickey, Y.Wu, and L.Malkas, Helicase
inhibition by anthracycline
anticancer agents, Mol.Pharmacol. 41(1992) 993-998.
57. R.Olinski, P.Jaruga, M.Foksinski, K.Bialkowski, and J.Tujakowski,
Epirubicin-induced oxidative DNA
damage and evidence for its repair in lymphocytes of cancer patients who are
undergoing
chemotherapy, Mol.Pharmacol. 52 (1997) 882-885.
58. U.Vaishampayan, R.E.Parchment, B.R.Jasti, and M.Hussain, Taxanes: an
overview of the
pharmacokinetics and pharmacodynamics, Urology 54 (1999) 22-29.
59. D.L.Morse, H.Gray, C.M.Payne, and R.J.Gillies, Docetaxel induces cell
death through mitotic
catastrophe in human breast cancer cells, Mol.Cancer Ther. 4 (2005) 1495-1504.
60. T.Wieder, F.Essmann, A.Prokop, K.Schmelz, K.Schulze-Osthoff, R.Beyaert,
B.Dorken, and
P.T.Daniel, Activation of caspase-8 in drug-induced apoptosis of B-lymphoid
cells is independent of
CD95/Fas receptor-ligand interaction and occurs downstream of caspase-3, Blood
97 (2001) 1378-
1387.
61. Mueller, 0. L., S.;Schroeder, A (2004) RNA Integrity Number (RIN) -
Standardization of RNA Quality Control.
62. Agilent 2100 Bioanalyzer,Agilent Technologies, Inc.2100 Expert User's
Guide. Agilent Technologies
Hewlett-Packard Str. 876337 Waldbronn Germany, Agilent Technologies, Inc. .
Agilent2100 G2946-
90004_Vespucci_UG_eBook_(NoSecPack)[2].
63. F Xia and SN Powell, 2002. "The molecular basis of radiosensitivity
and chemosensitivity in the
treatment of breast cancer". Semin Radiat Oncol (2002), 12(4) 296-304.
64. Hu, Z-P et al., 2012. "Metabolomic response of human skin tissue to low
dose ionizing radiation". Mol
BioSyst 8; 1979-1986.
65. Delic, J. et al., 1993. "Gamma-ray induced transcription and apoptosis-
associated loss of 28S rRNA
in interphase human lymphocytes". Int J Radiat Biol 64; 39-46.
66. Al-Mayah, A.H.J. et al., 2012. "Possible role of exosomes containing
RNA in mediating nontargeted
effect of ionizing radiation". Radiat Research 177; 539-545.
67 Krolak, J.M. et al., 1989. "18S Ribosomal RNA is Degraded during
Ribosome Maturation in Irradiated
HeLa cells". Radiat Research 118; 330-340.
51
7944618
Date Recue/Date Received 2022-10-28